Community differentiation of the cutaneous microbiota in psoriasis
- PMID: 24451201
- PMCID: PMC4177411
- DOI: 10.1186/2049-2618-1-31
Community differentiation of the cutaneous microbiota in psoriasis
Abstract
Background: Psoriasis is a common chronic inflammatory disease of the skin. We sought to characterize and compare the cutaneous microbiota of psoriatic lesions (lesion group), unaffected contralateral skin from psoriatic patients (unaffected group), and similar skin loci in matched healthy controls (control group) in order to discern patterns that govern skin colonization and their relationship to clinical diagnosis.
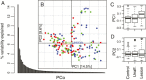
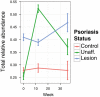
Results: Using high-throughput 16S rRNA gene sequencing, we assayed the cutaneous bacterial communities of 51 matched triplets and characterized these samples using community data analysis techniques. Intragroup Unifrac β diversity revealed increasing diversity from control to unaffected to lesion specimens. Likewise, principal coordinates analysis (PCoA) revealed separation of the lesion samples from unaffected and control along the first axis, suggesting that psoriasis is a major contributor to the observed diversity. The taxonomic richness and evenness decreased in both lesion and unaffected communities compared to control. These differences are explained by the combined increased abundance of the four major skin-associated genera (Corynebacterium, Propionibacterium, Staphylococcus, and Streptococcus), which present a potentially useful predictor for clinical skin type. Psoriasis samples also showed significant univariate decreases in relative abundances and strong classification performance of Cupriavidus, Flavisolibacter, Methylobacterium, and Schlegelella genera versus controls. The cutaneous microbiota separated into two distinct clusters, which we call cutaneotypes: (1) Proteobacteria-associated microbiota, and (2) Firmicutes-associated and Actinobacteria-associated microbiota. Cutaneotype 2 is enriched in lesion specimens compared to control (odds ratio 3.52 (95% CI 1.44 to 8.98), P <0.01).
Conclusions: Our results indicate that psoriasis induces physiological changes both at the lesion site and at the systemic level, which select for specific differential microbiota among the assayed clinical skin types. These differences in microbial community structure in psoriasis patients are potentially of pathophysiologic and diagnostic significance.
Figures







References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

