Characterization of all family-9 glycoside hydrolases synthesized by the cellulosome-producing bacterium Clostridium cellulolyticum
- PMID: 24451379
- PMCID: PMC3953250
- DOI: 10.1074/jbc.M113.545046
Characterization of all family-9 glycoside hydrolases synthesized by the cellulosome-producing bacterium Clostridium cellulolyticum
Abstract
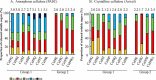
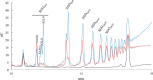
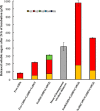
The genome of Clostridium cellulolyticum encodes 13 GH9 enzymes that display seven distinct domain organizations. All but one contain a dockerin module and were formerly detected in the cellulosomes, but only three of them were previously studied (Cel9E, Cel9G, and Cel9M). In this study, the 10 uncharacterized GH9 enzymes were overproduced in Escherichia coli and purified, and their activity pattern was investigated in the free state or in cellulosome chimeras with key cellulosomal cellulases. The newly purified GH9 enzymes, including those that share similar organization, all exhibited distinct activity patterns, various binding capacities on cellulosic substrates, and different synergies with pivotal cellulases in mini-cellulosomes. Furthermore, one enzyme (Cel9X) was characterized as the first genuine endoxyloglucanase belonging to this family, with no activity on soluble and insoluble celluloses. Another GH9 enzyme (Cel9V), whose sequence is 78% identical to the cellulosomal cellulase Cel9E, was found inactive in the free and complexed states on all tested substrates. The sole noncellulosomal GH9 (Cel9W) is a cellulase displaying a broad substrate specificity, whose engineered form bearing a dockerin can act synergistically in minicomplexes. Finally, incorporation of all GH9 cellulases in trivalent cellulosome chimera containing Cel48F and Cel9G generated a mixture of heterogeneous mini-cellulosomes that exhibit more activity on crystalline cellulose than the best homogeneous tri-functional complex. Altogether, our data emphasize the importance of GH9 diversity in bacterial cellulosomes, confirm that Cel9G is the most synergistic GH9 with the major endoprocessive cellulase Cel48F, but also identify Cel9U as an important cellulosomal component during cellulose depolymerization.
Keywords: Carbohydrate; Cellulase; Cellulose; Cellulosome; Clostridium cellulolyticum; Enzyme Kinetics; Enzymes; GH9; Protein Complexes; Synergy.
Figures




References
-
- Henrissat B., Claeyssens M., Tomme P., Lemesle L., Mornon J. P. (1989) Cellulase families revealed by hydrophobic cluster analysis. Gene 81, 83–95 - PubMed
-
- Honda Y., Shimaya N., Ishisaki K., Ebihara M., Taniguchi H. (2011) Elucidation of exo-β-d-glucosaminidase activity of a family 9 glycoside hydrolase (PBPRA0520) from Photobacterium profundum SS9. Glycobiology 21, 503–511 - PubMed
-
- Arai T., Araki R., Tanaka A., Karita S., Kimura T., Sakka K., Ohmiya K. (2003) Characterization of a cellulase containing a family 30 carbohydrate-binding module (CBM) derived from Clostridium thermocellum CelJ: importance of the CBM to cellulose hydrolysis. J. Bacteriol. 185, 504–512 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

