FOXL2 transcriptionally represses Sf1 expression by antagonizing WT1 during ovarian development in mice
- PMID: 24451388
- PMCID: PMC3986836
- DOI: 10.1096/fj.13-246108
FOXL2 transcriptionally represses Sf1 expression by antagonizing WT1 during ovarian development in mice
Abstract
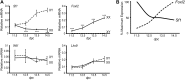
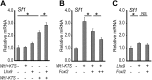
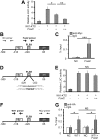
Steroidogenic factor 1 (SF1; Ad4BP/NR5A1) plays key roles in gonadal development. Initially, the Sf1 gene is expressed in mouse fetal gonads of both sexes, but later is up-regulated in testes and down-regulated in ovaries. While Sf1 expression is activated and maintained by Wilms tumor 1 (WT1) and LIM homeobox 9 (LHX9), the mechanism of sex-specific regulation remains unclear. We hypothesized that Sf1 is repressed by the transcription factor Forkhead box L2 (FOXL2) during ovarian development. In an in vitro system (TM3 cells), up-regulation of Sf1 by the WT1 splice variant WT1-KTS was antagonized by FOXL2, as determined by quantitative RT-PCR. Using reporter assays, we localized the Sf1 proximal promoter region involved in this antagonism to a 674-bp interval. A conserved FOXL2 binding site was identified in this interval by in vitro chromatin immunoprecipitation. Introducing mutations into this site abolished negative regulation by FOXL2 in reporter assays. Finally, in Foxl2-null mice, Sf1 expression was increased 2-fold relative to wild-type XX fetal gonads. Our results support the hypothesis that FOXL2 negatively regulates Sf1 expression by antagonizing WT1-KTS during early ovarian development in mice.
Keywords: LHX9; reproduction; sex determination.
Figures

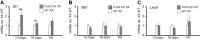
Similar articles
-
Antagonistic regulation of Cyp26b1 by transcription factors SOX9/SF1 and FOXL2 during gonadal development in mice.FASEB J. 2011 Oct;25(10):3561-9. doi: 10.1096/fj.11-184333. Epub 2011 Jul 14. FASEB J. 2011. PMID: 21757499 Free PMC article.
-
The Wilms tumor suppressor WT1 regulates early gonad development by activation of Sf1.Genes Dev. 2002 Jul 15;16(14):1839-51. doi: 10.1101/gad.220102. Genes Dev. 2002. PMID: 12130543 Free PMC article.
-
Expression of inhibin-alpha is regulated synergistically by Wilms' tumor gene 1 (Wt1) and steroidogenic factor-1 (Sf1) in sertoli cells.PLoS One. 2013;8(1):e53140. doi: 10.1371/journal.pone.0053140. Epub 2013 Jan 11. PLoS One. 2013. PMID: 23326390 Free PMC article.
-
Determination and stability of gonadal sex.J Androl. 2010 Jan-Feb;31(1):16-25. doi: 10.2164/jandrol.109.008201. Epub 2009 Oct 29. J Androl. 2010. PMID: 19875493 Free PMC article. Review.
-
Early gonadal development: exploring Wt1 and Sox9 function.Novartis Found Symp. 2002;244:23-31; discussion 31-42, 253-7. Novartis Found Symp. 2002. PMID: 11990794 Review.
Cited by
-
Peptidyl arginine deiminase 2 (Padi2) is expressed in Sertoli cells in a specific manner and regulated by SOX9 during testicular development.Sci Rep. 2018 Sep 5;8(1):13263. doi: 10.1038/s41598-018-31376-8. Sci Rep. 2018. PMID: 30185873 Free PMC article.
-
The p.R92W variant of NR5A1/Nr5a1 induces testicular development of 46,XX gonads in humans, but not in mice: phenotypic comparison of human patients and mutation-induced mice.Biol Sex Differ. 2016 Nov 8;7:56. doi: 10.1186/s13293-016-0114-6. eCollection 2016. Biol Sex Differ. 2016. PMID: 27833742 Free PMC article.
-
Genetics of the ovarian reserve.Front Genet. 2015 Oct 15;6:308. doi: 10.3389/fgene.2015.00308. eCollection 2015. Front Genet. 2015. PMID: 26528328 Free PMC article. Review.
-
MicroRNA-764-3p regulates 17β-estradiol synthesis of mouse ovarian granulosa cells by targeting steroidogenic factor-1.In Vitro Cell Dev Biol Anim. 2016 Mar;52(3):365-373. doi: 10.1007/s11626-015-9977-9. Epub 2015 Dec 16. In Vitro Cell Dev Biol Anim. 2016. PMID: 26676955
-
Ex vivo cultures combined with vivo-morpholino induced gene knockdown provide a system to assess the role of WT1 and GATA4 during gonad differentiation.PLoS One. 2017 Apr 20;12(4):e0176296. doi: 10.1371/journal.pone.0176296. eCollection 2017. PLoS One. 2017. PMID: 28426816 Free PMC article.
References
-
- Morohashi K., Honda S., Inomata Y., Handa H., Omura T. (1992) A common trans-acting factor, Ad4-binding protein, to the promoters of steroidogenic P-450s. J. Biol. Chem. 267, 17913–17919 - PubMed
-
- Rice D. A., Mouw A. R., Bogerd A. M., Parker K. L. (1991) A shared promoter element regulates the expression of three steridogenic enzymes. Mol. Endocrinol. 5, 1552–1561 - PubMed
-
- Ikeda Y., Shen W. H., Ingraham H. A., Parker K. L. (1994) Developmental expression of mouse steroidogenic factor-1, an essential regulator of the steroid hydroxylases. Mol. Endocrinol. 8, 654–662 - PubMed
-
- Ingraham H. A., Lala D. S., Ikeda Y., Luo X., Shen W. H., Nachtigal M. W., Abbud R., Nilson J. H., Parker K. L. (1994) The nuclear receptor steroidogenic factor 1 acts at multiple levels of the reproductive axis. Genes Dev. 8, 2302–2312 - PubMed
-
- Luo X., Ikeda Y., Parker K. L. (1994) A cell-specific nuclear receptor is essential for adrenal and gonadal development and sexual differentiation. Cell 77, 481–490 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

