A well-constrained estimate for the timing of the salmonid whole genome duplication reveals major decoupling from species diversification
- PMID: 24452024
- PMCID: PMC3906940
- DOI: 10.1098/rspb.2013.2881
A well-constrained estimate for the timing of the salmonid whole genome duplication reveals major decoupling from species diversification
Abstract
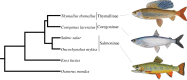
Whole genome duplication (WGD) is often considered to be mechanistically associated with species diversification. Such ideas have been anecdotally attached to a WGD at the stem of the salmonid fish family, but remain untested. Here, we characterized an extensive set of gene paralogues retained from the salmonid WGD, in species covering the major lineages (subfamilies Salmoninae, Thymallinae and Coregoninae). By combining the data in calibrated relaxed molecular clock analyses, we provide the first well-constrained and direct estimate for the timing of the salmonid WGD. Our results suggest that the event occurred no later in time than 88 Ma and that 40-50 Myr passed subsequently until the subfamilies diverged. We also recovered a Thymallinae-Coregoninae sister relationship with maximal support. Comparative phylogenetic tests demonstrated that salmonid diversification patterns are closely allied in time with the continuous climatic cooling that followed the Eocene-Oligocene transition, with the highest diversification rates coinciding with recent ice ages. Further tests revealed considerably higher speciation rates in lineages that evolved anadromy--the physiological capacity to migrate between fresh and seawater--than in sister groups that retained the ancestral state of freshwater residency. Anadromy, which probably evolved in response to climatic cooling, is an established catalyst of genetic isolation, particularly during environmental perturbations (for example, glaciation cycles). We thus conclude that climate-linked ecophysiological factors, rather than WGD, were the primary drivers of salmonid diversification.
Keywords: anadromy; climate change; evolution; salmonid fish; species diversification; whole genome duplication.
Figures

References
-
- Lynch M. 2007. The origins of genome architecture, 1st edn Sunderland, MA: Sinauer Associates
-
- Van de Peer Y, Maere S, Meyer A. 2009. The evolutionary significance of ancient genome duplication. Nat. Rev. Genet. 10, 725–732 (doi:10.1038/nrg2600) - DOI - PubMed
-
- Scannell DR, Byrne KP, Gordon JL, Wong S, Wolfe KH. 2006. Multiple rounds of speciation associated with reciprocal gene loss in polyploid yeasts. Nature 440, 341–345 (doi:10.1038/nature04562) - DOI - PubMed
-
- Jiao Y, et al. 2011. Ancestral polyploidy in seed plants and angiosperms. Nature 473, 97–100 (doi:10.1038/nature0991) - DOI - PubMed
-
- Soltis DE, et al. 2009. Polyploidy and angiosperm diversification. Am. J. Bot. 96, 336–348 (doi:10.3732/ajb.0800079) - DOI - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
