Characterization of cleavage intermediate and star sites of RM.Tth111II
- PMID: 24452415
- PMCID: PMC3899748
- DOI: 10.1038/srep03838
Characterization of cleavage intermediate and star sites of RM.Tth111II
Abstract
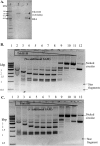
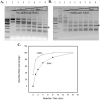
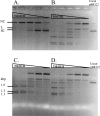
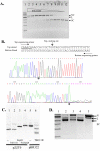
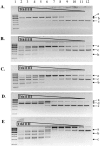
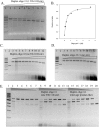
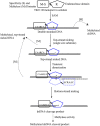
Tth111II is a thermostable Type IIGS restriction enzyme that recognizes DNA sites CAARCA (R = A or G) and cleaves downstream at N11/N9. Here, the tth111IIRM gene was cloned and expressed in E. coli, and Tth111II was purified. The purified enzyme contains internally-bound S-adenosylmethionine (SAM). When the internal SAM was removed, the endonuclease activity was stimulated by adding SAM or its analog sinefungin. The cleavage intermediate is mostly top-strand nicked DNA on a single-site plasmid. Addition of duplex oligos with a cognate site stimulates cleavage activity of the one-site substrate. Tth111II cleaves a two-site plasmid DNA with equal efficiency regardless of site orientation. We propose the top-strand nicking is carried out by a Tth111II monomer and bottom-strand cleavage is carried out by a transient dimer. Tth111II methylates cleavage product-like duplex oligos CAAACAN9, but the modification rate is estimated to be much slower than the top-strand nicking rate. We cloned and sequenced a number of Tth111II star sites which are 1-bp different from the cognate sites. A biochemical pathway is proposed for the restriction and methylation activities of Tth111II.
Figures

References
-
- Shapovalova N. I., Zheleznaia L. A., Matvienko N. N. & Matvienko N. I. [A new site-specific endonuclease BspKT51]. Biokhimiia 59, 485–493 (1994). - PubMed
-
- Janulaitis A., Bitinaite J. & Jaskeleviciene B. A new sequence-specific endonuclease from Gluconobacter suboxydans. FEBS Lett 151, 243–247 (1983). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

