A day in the life of the spliceosome
- PMID: 24452469
- PMCID: PMC4060434
- DOI: 10.1038/nrm3742
A day in the life of the spliceosome
Erratum in
- Nat Rev Mol Cell Biol. 2014 Apr;15(4):294
Abstract
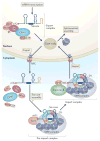
One of the most amazing findings in molecular biology was the discovery that eukaryotic genes are discontinuous, with coding DNA being interrupted by stretches of non-coding sequence. The subsequent realization that the intervening regions are removed from pre-mRNA transcripts via the activity of a common set of small nuclear RNAs (snRNAs), which assemble together with associated proteins into a complex known as the spliceosome, was equally surprising. How do cells coordinate the assembly of this molecular machine? And how does the spliceosome accurately recognize exons and introns to carry out the splicing reaction? Insights into these questions have been gained by studying the life cycle of spliceosomal snRNAs from their transcription, nuclear export and re-import to their dynamic assembly into the spliceosome. This assembly process can also affect the regulation of alternative splicing and has implications for human disease.
Figures



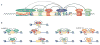
References
-
- Chow LT, Gelinas RE, Broker TR, Roberts RJ. An amazing sequence arrangement at the 5′ ends of adenovirus 2 messenger RNA. Cell. 1977;12:1–8. - PubMed
-
- Lerner MR, Boyle JA, Mount SM, Wolin SL, Steitz JA. Are snRNPs involved in splicing? Nature. 1980;283:220–4. - PubMed
-
- Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Molecular cell. 2003;12:5–14. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

