Regulated protein turnover: snapshots of the proteasome in action
- PMID: 24452470
- PMCID: PMC4384331
- DOI: 10.1038/nrm3741
Regulated protein turnover: snapshots of the proteasome in action
Abstract
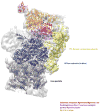
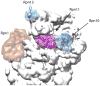
The ubiquitin proteasome system (UPS) is the main ATP-dependent protein degradation pathway in the cytosol and nucleus of eukaryotic cells. At its centre is the 26S proteasome, which degrades regulatory proteins and misfolded or damaged proteins. In a major breakthrough, several groups have determined high-resolution structures of the entire 26S proteasome particle in different nucleotide conditions and with and without substrate using cryo-electron microscopy combined with other techniques. These structures provide some surprising insights into the functional mechanism of the proteasome and will give invaluable guidance for genetic and biochemical studies of this key regulatory system.
Figures
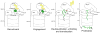



References
-
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–79. - PubMed
-
- Varshavsky A. The ubiquitin system. Trends Biochem Sci. 1997;22:383–7. - PubMed
-
- Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–47. - PubMed
-
- Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–39. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

