Molecular profiling of chordoma
- PMID: 24452533
- PMCID: PMC3977807
- DOI: 10.3892/ijo.2014.2268
Molecular profiling of chordoma
Abstract
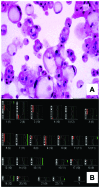
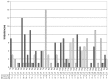
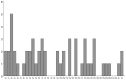
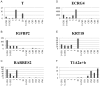
The molecular basis of chordoma is still poorly understood, particularly with respect to differentially expressed genes involved in the primary origin of chordoma. In this study, therefore, we compared the transcriptional expression profile of one sacral chordoma recurrence, two chordoma cell lines (U-CH1 and U-CH2) and one chondrosarcoma cell line (U-CS2) with vertebral disc using a high-density oligonucleotide array. The expression of 65 genes whose mRNA levels differed significantly (p<0.001; ≥6-fold change) between chordoma and control (vertebral disc) was identified. Genes with increased expression in chordoma compared to control and chondrosarcoma were most frequently located on chromosomes 2 (11%), 5 (8%), 1 and 7 (each 6%), whereas interphase cytogenetics of 33 chordomas demonstrated gains of chromosomal material most prevalent on 7q (42%), 12q (21%), 17q (21%), 20q (27%) and 22q (21%). The microarray data were confirmed for selected genes by quantitative polymerase chain reaction analysis. As in other studies, we showed the expression of brachyury. We demonstrate the expression of new potential candidates for chordoma tumorigenesis, such as CD24, ECRG4, RARRES2, IGFBP2, RAP1, HAI2, RAB38, osteopontin, GalNAc-T3, VAMP8 and others. Thus, we identified and validated a set of interesting candidate genes whose differential expression likely plays a role in chordoma.
Figures

References
-
- Dorfman HD, Czerniak B, editors. Bone tumors. Mosby; St. Louis, MO: 1998.
-
- Mirra JM, Nelson SD, Della Rocca C, Mertens F. Chordoma. In: Fletcher CDM, Unni KK, Mertens F, editors. Pathology and Genetics of Tumors of Soft Tissue and Bone World Health Organization Classification of Tumors. IARC Press; Lyon: 2002. pp. 316–317.
-
- Shalaby AA, Presneau N, Idowu BD, Thompson L, Briggs TR, Tirabosco R, Diss TC, Flanagan AM. Analysis of the fibroblastic growth factor receptor-RAS/RAF/MEK/ERK-ETS2/ brachyury signalling pathway in chordomas. Mod Pathol. 2009;22:996–1005. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

