High resolution non-invasive detection of a fetal microdeletion using the GCREM algorithm
- PMID: 24452987
- PMCID: PMC4266320
- DOI: 10.1002/pd.4331
High resolution non-invasive detection of a fetal microdeletion using the GCREM algorithm
Abstract
Background/objective: The non-invasive prenatal detection of fetal microdeletions becomes increasingly challenging as the size of the mutation decreases, with current practical lower limits in the range of a few megabases. Our goals were to explore the lower limits of microdeletion size detection via non-invasive prenatal tests using Minimally Invasive Karyotyping (MINK) and introduce/evaluate a novel statistical approach we recently developed called the GC Content Random Effect Model (GCREM).
Methods: Maternal plasma was obtained from a pregnancy affected by a 4.2-Mb fetal microdeletion and three normal controls. Plasma DNA was subjected to capture an 8-Mb sequence spanning the breakpoint region and sequence. Data were analyzed with our published method, MINK, and a new method called GCREM.
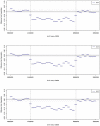
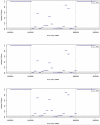
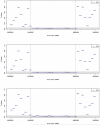
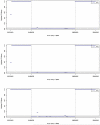
Results: The 8-Mb capture segment was divided into either 38 or 76 non-overlapping regions of 200 and 100 Kb, respectively. At 200 Kb resolution, using GCREM (but not MINK), we obtained significant adjusted p-values for all 20 regions overlapping the deleted sequence, and non-significant p-values for all 18 reference regions. At 100 Kb resolution, GCREM identified significant adjusted p-values for all but one 100-Kb region located inside the deleted region.
Conclusion: Targeted sequencing and GCREM analysis may enable cost effective detection of fetal microdeletions and microduplications at high resolution.
© 2014 John Wiley & Sons, Ltd.
Figures

Similar articles
-
Comparative evaluation of the Minimally-Invasive Karyotyping (MINK) algorithm for non-invasive prenatal testing.PLoS One. 2017 Mar 17;12(3):e0171882. doi: 10.1371/journal.pone.0171882. eCollection 2017. PLoS One. 2017. PMID: 28306738 Free PMC article.
-
A method for noninvasive detection of fetal large deletions/duplications by low coverage massively parallel sequencing.Prenat Diagn. 2013 Jun;33(6):584-90. doi: 10.1002/pd.4110. Prenat Diagn. 2013. PMID: 23592436
-
Noninvasive prenatal molecular karyotyping from maternal plasma.PLoS One. 2013 Apr 17;8(4):e60968. doi: 10.1371/journal.pone.0060968. Print 2013. PLoS One. 2013. PMID: 23613765 Free PMC article.
-
Non-invasive prenatal diagnostics of aneuploidy using next-generation DNA sequencing technologies, and clinical considerations.Clin Chem Lab Med. 2013 Jun;51(6):1141-54. doi: 10.1515/cclm-2012-0281. Clin Chem Lab Med. 2013. PMID: 23023923 Review.
-
Non-invasive prenatal testing using massively parallel sequencing of maternal plasma DNA: from molecular karyotyping to fetal whole-genome sequencing.Reprod Biomed Online. 2013 Dec;27(6):593-8. doi: 10.1016/j.rbmo.2013.08.008. Epub 2013 Sep 7. Reprod Biomed Online. 2013. PMID: 24140310 Review.
Cited by
-
High Levels of Sample-to-Sample Variation Confound Data Analysis for Non-Invasive Prenatal Screening of Fetal Microdeletions.PLoS One. 2016 Jun 1;11(6):e0153182. doi: 10.1371/journal.pone.0153182. eCollection 2016. PLoS One. 2016. PMID: 27249650 Free PMC article.
-
Recent advances of genomic testing in perinatal medicine.Semin Perinatol. 2015 Feb;39(1):44-54. doi: 10.1053/j.semperi.2014.10.009. Epub 2014 Nov 28. Semin Perinatol. 2015. PMID: 25444417 Free PMC article. Review.
-
Comprehensive Evaluation of Non-invasive Prenatal Screening to Detect Fetal Copy Number Variations.Front Genet. 2021 Jul 16;12:665589. doi: 10.3389/fgene.2021.665589. eCollection 2021. Front Genet. 2021. PMID: 34335682 Free PMC article.
-
Optimization of techniques for multiple platform testing in small, precious samples such as human chorionic villus sampling.Prenat Diagn. 2016 Nov;36(11):1061-1070. doi: 10.1002/pd.4936. Epub 2016 Nov 7. Prenat Diagn. 2016. PMID: 27718505 Free PMC article.
-
Noninvasive prenatal testing using a novel analysis pipeline to screen for all autosomal fetal aneuploidies improves pregnancy management.Eur J Hum Genet. 2015 Oct;23(10):1286-93. doi: 10.1038/ejhg.2014.282. Epub 2015 Jan 14. Eur J Hum Genet. 2015. PMID: 25585704 Free PMC article.
References
-
- Hertling-Schaal E, Perrotin F, de Poncheville L, et al. Maternal anxiety induced by prenatal diagnostic techniques: detection and management. Gynecol Obstet Fertil. 2001 Jun;29(6):440–6. - PubMed
-
- Hewison J, Nixon J, Fountain J, et al. Amniocentesis results: Investigation of anxiety. The ARIA trial. Health Technol Assess. 2006 Dec;10(50):iii. ix-x, 1-78. - PubMed
-
- Hewison J, Nixon J, Fountain J, et al. A randomised trial of two methods of issuing prenatal test results: the ARIA (Amniocentesis Results: Investigation of Anxiety) trial. BJOG. 2007 Apr;114(4):462–8. - PubMed
-
- Mujezinovic F, Alfirevic Z. Procedure-related complications of amniocentesis and chorionic villous sampling: a systematic review. Obstet Gynecol. 2007 Sep;110(3):687–94. - PubMed
-
- Odibo AO, Gray DL, Dicke JM, et al. Revisiting the fetal loss rate after second-trimester genetic amniocentesis: a single center's 16-year experience. Obstet Gynecol. 2008 Mar;111(3):589–95. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

