Consumption of acidic water alters the gut microbiome and decreases the risk of diabetes in NOD mice
- PMID: 24453191
- PMCID: PMC3966285
- DOI: 10.1369/0022155413519650
Consumption of acidic water alters the gut microbiome and decreases the risk of diabetes in NOD mice
Abstract
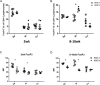
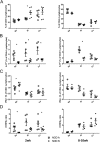
Infant formula and breastfeeding are environmental factors that influence the incidence of Type 1 Diabetes (T1D) as well as the acidity of newborn diets. To determine if altering the intestinal microbiome is one mechanism through which an acidic liquid plays a role in T1D, we placed non-obese diabetic (NOD)/ShiLtJt mice on neutral (N) or acidified H2O and monitored the impact on microbial composition and diabetes incidence. NOD-N mice showed an increased development of diabetes, while exhibiting a decrease in Firmicutes and an increase in Bacteroidetes, Actinobacteria, and Proteobacteria from as early as 2 weeks of age. NOD-N mice had a decrease in the levels of Foxp3 expression in CD4(+)Foxp3(+) cells, as well as decreased CD4(+)IL17(+) cells, and a lower ratio of IL17/IFNγ CD4+ T-cells. Our data clearly indicates that a change in the acidity of liquids consumed dramatically alters the intestinal microbiome, the presence of protective Th17 and Treg cells, and the incidence of diabetes. This data suggests that early dietary manipulation of intestinal microbiota may be a novel mechanism to delay T1D onset in genetically pre-disposed individuals.
Keywords: T-cells; diet; microbiota; mouse model; type 1 diabetes.
Conflict of interest statement
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

