Expression of proteolipid protein gene in spinal cord stem cells and early oligodendrocyte progenitor cells is dispensable for normal cell migration and myelination
- PMID: 24453324
- PMCID: PMC3898293
- DOI: 10.1523/JNEUROSCI.2477-13.2014
Expression of proteolipid protein gene in spinal cord stem cells and early oligodendrocyte progenitor cells is dispensable for normal cell migration and myelination
Abstract
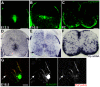
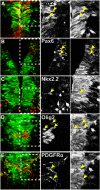
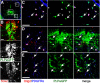
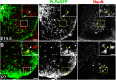
Plp1 gene expression occurs very early in development, well before the onset of myelination, creating a conundrum with regard to the function of myelin proteolipid protein (PLP), one of the major proteins in compact myelin. Using PLP-EGFP mice to investigate Plp1 promoter activity, we found that, at very early time points, PLP-EGFP was expressed in Sox2+ undifferentiated precursors in the spinal cord ventricular zone (VZ), as well as in the progenitors of both neuronal and glial lineages. As development progressed, most PLP-EGFP-expressing cells gave rise to oligodendrocyte progenitor cells (OPCs). The expression of PLP-EGFP in the spinal cord was quite dynamic during development. PLP-EGFP was highly expressed as cells delaminated from the VZ. Expression was downregulated as cells moved laterally through the cord, and then robustly upregulated as OPCs differentiated into mature myelinating oligodendrocytes. The presence of PLP-EGFP expression in OPCs raises the question of its role in this migratory population. We crossed PLP-EGFP reporter mice into a Plp1-null background to investigate the role of PLP in early OPC development. In the absence of PLP, normal numbers of OPCs were generated and their distribution throughout the spinal cord was unaffected. However, the orientation and length of OPC processes during migration was abnormal in Plp1-null mice, suggesting that PLP plays a role either in the structural integrity of OPC processes or in their response to extracellular cues that orient process outgrowth.
Keywords: OPC; PLP; migration; myelination; oligodendrocyte; proteolipid protein.
Figures



Similar articles
-
Myelin Proteolipid Protein Complexes with αv Integrin and AMPA Receptors In Vivo and Regulates AMPA-Dependent Oligodendrocyte Progenitor Cell Migration through the Modulation of Cell-Surface GluR2 Expression.J Neurosci. 2015 Aug 26;35(34):12018-32. doi: 10.1523/JNEUROSCI.5151-14.2015. J Neurosci. 2015. PMID: 26311781 Free PMC article.
-
Identity and lineage fate of proteolipid protein 1 gene (Plp1)-expressing cells in the embryonic murine spinal cord.Dev Dyn. 2020 Aug;249(8):946-960. doi: 10.1002/dvdy.184. Epub 2020 May 26. Dev Dyn. 2020. PMID: 32353175
-
Cyclin dependent kinase 5 is required for the normal development of oligodendrocytes and myelin formation.Dev Biol. 2013 Jun 15;378(2):94-106. doi: 10.1016/j.ydbio.2013.03.023. Epub 2013 Apr 10. Dev Biol. 2013. PMID: 23583582 Free PMC article.
-
The early steps of oligodendrogenesis: insights from the study of the plp lineage in the brain of chicks and rodents.Dev Neurosci. 2001;23(4-5):318-26. doi: 10.1159/000048715. Dev Neurosci. 2001. PMID: 11756747 Review.
-
Electrical signalling properties of oligodendrocyte precursor cells.Neuron Glia Biol. 2009 May;5(1-2):3-11. doi: 10.1017/S1740925X09990202. Epub 2009 Aug 13. Neuron Glia Biol. 2009. PMID: 19674511 Review.
Cited by
-
Oligodendrogenesis and myelination tracing in a CRISPR/Cas9-engineered brain microphysiological system.Front Cell Neurosci. 2023 Jan 19;16:1094291. doi: 10.3389/fncel.2022.1094291. eCollection 2022. Front Cell Neurosci. 2023. PMID: 36744062 Free PMC article.
-
The formation of a glial scar does not prohibit remyelination in an animal model of multiple sclerosis.Glia. 2019 Mar;67(3):467-481. doi: 10.1002/glia.23556. Epub 2018 Nov 28. Glia. 2019. PMID: 30484905 Free PMC article.
-
Exploring synergistic effects: Atorvastatin and electrical stimulation in spinal cord injury therapy.IBRO Neurosci Rep. 2025 Feb 25;18:389-399. doi: 10.1016/j.ibneur.2025.02.012. eCollection 2025 Jun. IBRO Neurosci Rep. 2025. PMID: 40124115 Free PMC article.
-
Development of myelinating glia: An overview.Glia. 2022 Dec;70(12):2237-2259. doi: 10.1002/glia.24238. Epub 2022 Jul 4. Glia. 2022. PMID: 35785432 Free PMC article. Review.
-
Insights into olfactory ensheathing cell development from a laser-microdissection and transcriptome-profiling approach.Glia. 2020 Dec;68(12):2550-2584. doi: 10.1002/glia.23870. Epub 2020 Aug 28. Glia. 2020. PMID: 32857879 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
