Identification of proteins that interact with alpha A-crystallin using a human proteome microarray
- PMID: 24453475
- PMCID: PMC3893783
Identification of proteins that interact with alpha A-crystallin using a human proteome microarray
Abstract
Purpose: To identify proteins interacting with alpha A-crystallin (CRYAA) and to investigate the potential role that these protein interactions play in the function of CRYAA using a human proteome (HuProt) microarray.
Methods: The active full-length CRYAA protein corresponding to amino acids 1-173 of CRYAA was recombined. A HuProt microarray composed of 17,225 human full-length proteins with N-terminal glutathione S-transferase (GST) tags was used to identify protein-protein interactions. The probes were considered detectable when the signal to noise ratio (SNR) was over 1.2. The identified proteins were subjected to subsequent bioinformatics analysis using the DAVID database.
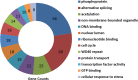
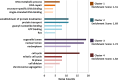
Results: The HuProt microarray results showed that the signals of 343 proteins were higher in the recombinant CRYAA group than in the control group. The SNR of 127 proteins was ≥ 1.2. The SNR of the following eight proteins was > 3.0: hematopoietic cell-specific Lyn substrate 1 (HCLS1), Kelch domain-containing 6 (KLHDC6), sarcoglycan delta (SGCD), KIAA1706 protein (KIAA1706), RNA guanylyltransferase and 5'-phosphatase (RNGTT), chromosome 10 open reading frame 57 (C10orf57), chromosome 9 open reading frame 52 (C9orf52), and plasminogen activator, urokinase receptor (PLAUR). The bioinformatics analysis revealed 127 proteins associated with phosphoproteins, alternative splicing, acetylation, DNA binding, the nuclear lumen, ribonucleotide binding, the cell cycle, WD40 repeats, protein transport, transcription factor activity, GTP binding, and cellular response to stress. Functional annotation clustering showed that they belong to cell cycle, organelle or nuclear lumen, protein transport, and DNA binding and repair clusters. CRYAA interacted with these proteins to maintain their solubility and decrease the accumulation of denatured target proteins. The protein-protein interactions may help CRYAA carry out multifaceted functions.
Conclusions: One-hundred and twenty-seven of 17,225 human full-length proteins were identified that interact with CRYAA. The advent of microarray analysis enables a better understanding of the functions of CRYAA as a molecular chaperone.
Figures

References
-
- Kato K, Shinohara H, Kurobe N, Goto S, Inaguma Y, Ohshima K. Immunoreactive alpha A crystallin in rat non-lenticular tissues detected with a sensitive immunoassay method. Biochim Biophys Acta. 1991;1080:173–80. - PubMed
-
- Srinivasan AN, Nagineni CN, Bhat SP. alpha A-crystallin is expressed in non-ocular tissues. J Biol Chem. 1992;267:23337–41. - PubMed
-
- Zhou P, Luo Y, Liu X, Fan L, Lu Y. Down-regulation and CpG island hypermethylation of CRYAA in age-related nuclear cataract. FASEB J. 2012;26:4897–902. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

