Different levels of catabolite repression optimize growth in stable and variable environments
- PMID: 24453942
- PMCID: PMC3891604
- DOI: 10.1371/journal.pbio.1001764
Different levels of catabolite repression optimize growth in stable and variable environments
Abstract
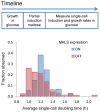
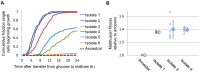
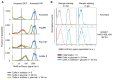
Organisms respond to environmental changes by adapting the expression of key genes. However, such transcriptional reprogramming requires time and energy, and may also leave the organism ill-adapted when the original environment returns. Here, we study the dynamics of transcriptional reprogramming and fitness in the model eukaryote Saccharomyces cerevisiae in response to changing carbon environments. Population and single-cell analyses reveal that some wild yeast strains rapidly and uniformly adapt gene expression and growth to changing carbon sources, whereas other strains respond more slowly, resulting in long periods of slow growth (the so-called "lag phase") and large differences between individual cells within the population. We exploit this natural heterogeneity to evolve a set of mutants that demonstrate how the frequency and duration of changes in carbon source can favor different carbon catabolite repression strategies. At one end of this spectrum are "specialist" strategies that display high rates of growth in stable environments, with more stringent catabolite repression and slower transcriptional reprogramming. The other mutants display less stringent catabolite repression, resulting in leaky expression of genes that are not required for growth in glucose. This "generalist" strategy reduces fitness in glucose, but allows faster transcriptional reprogramming and shorter lag phases when the cells need to shift to alternative carbon sources. Whole-genome sequencing of these mutants reveals that mutations in key regulatory genes such as HXK2 and STD1 adjust the regulation and transcriptional noise of metabolic genes, with some mutations leading to alternative gene regulatory strategies that allow "stochastic sensing" of the environment. Together, our study unmasks how variable and stable environments favor distinct strategies of transcriptional reprogramming and growth.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Kussell E, Leibler S (2005) Phenotypic diversity, population growth, and information in fluctuating environments. Science 309: 2075–2078 doi:10.1126/science.1114383 - DOI - PubMed
-
- Stephens DW, Krebs JR (1986) Foraging theory. Princeton, NJ: Princeton University Press.
-
- Dekel E, Alon U (2005) Optimality and evolutionary tuning of the expression level of a protein. Nature 436: 588–592 doi:10.1038/nature03842 - DOI - PubMed
-
- Shoval O, Sheftel H, Shinar G, Hart Y, Ramote O, et al. (2012) Evolutionary trade-offs, pareto optimality, and the geometry of phenotype space. Science 336: 1157–1160 doi:10.1126/science.1217405 - DOI - PubMed
-
- Cooper VS, Lenski RE (2000) The population genetics of ecological specialization in evolving Escherichia coli populations. Nature 407: 736–739 doi:10.1038/35037572 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

