PEX5 and ubiquitin dynamics on mammalian peroxisome membranes
- PMID: 24453954
- PMCID: PMC3894153
- DOI: 10.1371/journal.pcbi.1003426
PEX5 and ubiquitin dynamics on mammalian peroxisome membranes
Abstract
Peroxisomes are membrane-bound organelles within eukaryotic cells that post-translationally import folded proteins into their matrix. Matrix protein import requires a shuttle receptor protein, usually PEX5, that cycles through docking with the peroxisomal membrane, ubiquitination, and export back into the cytosol followed by deubiquitination. Matrix proteins associate with PEX5 in the cytosol and are translocated into the peroxisome lumen during the PEX5 cycle. This cargo translocation step is not well understood, and its energetics remain controversial. We use stochastic computational models to explore different ways the AAA ATPase driven removal of PEX5 may couple with cargo translocation in peroxisomal importers of mammalian cells. The first model considered is uncoupled, in which translocation is spontaneous, and does not immediately depend on PEX5 removal. The second is directly coupled, in which cargo translocation only occurs when its PEX5 is removed from the peroxisomal membrane. The third, novel, model is cooperatively coupled and requires two PEX5 on a given importomer for cargo translocation--one PEX5 with associated cargo and one with ubiquitin. We measure both the PEX5 and the ubiquitin levels on the peroxisomes as we vary the matrix protein cargo addition rate into the cytosol. We find that both uncoupled and directly coupled translocation behave identically with respect to PEX5 and ubiquitin, and the peroxisomal ubiquitin signal increases as the matrix protein traffic increases. In contrast, cooperatively coupled translocation behaves dramatically differently, with a ubiquitin signal that decreases with increasing matrix protein traffic. Recent work has shown that ubiquitin on mammalian peroxisome membranes can lead to selective degradation by autophagy, or 'pexophagy.' Therefore, the high ubiquitin level for low matrix cargo traffic with cooperatively coupled protein translocation could be used as a disuse signal to mediate pexophagy. This mechanism may be one way that cells could regulate peroxisome numbers.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

 . There are
. There are  binding sites per importomer; here we illustrate
binding sites per importomer; here we illustrate  . (B) If unoccupied, the RING complex site is immediately occupied by another PEX5 on the importomer. (C) The RING complex (purple rectangle) will ubiquitinate an associated PEX5 at rate
. (B) If unoccupied, the RING complex site is immediately occupied by another PEX5 on the importomer. (C) The RING complex (purple rectangle) will ubiquitinate an associated PEX5 at rate  . We generally allow only one ubiquitinated PEX5 per importomer. For (A), (B), and (C) the AAA complex is shown, and will participate in PEX5 export as described in Fig. 2.
. We generally allow only one ubiquitinated PEX5 per importomer. For (A), (B), and (C) the AAA complex is shown, and will participate in PEX5 export as described in Fig. 2.
 . In uncoupled translocation, associated cargo is translocated spontaneously after binding to the importomer. (B) If translocation is uncoupled, then export of ubiquitinated PEX5 by the AAA complex at rate
. In uncoupled translocation, associated cargo is translocated spontaneously after binding to the importomer. (B) If translocation is uncoupled, then export of ubiquitinated PEX5 by the AAA complex at rate  does not have a relationship with cargo translocation. (C) In directly coupled translocation, the cargo translocation occurs as the ubiquitinated PEX5 is removed from the importomer by the AAA complex at rate
does not have a relationship with cargo translocation. (C) In directly coupled translocation, the cargo translocation occurs as the ubiquitinated PEX5 is removed from the importomer by the AAA complex at rate  . The PEX5 is shown simultaneously both cargo-loaded and ubiquitinated — this figure is meant to be illustrative; see Methods for discussion. (D) In cooperatively coupled translocation, the removal of PEX5 by the AAA complex (
. The PEX5 is shown simultaneously both cargo-loaded and ubiquitinated — this figure is meant to be illustrative; see Methods for discussion. (D) In cooperatively coupled translocation, the removal of PEX5 by the AAA complex ( ) can only occur when coupled to the cargo translocation of a distinct PEX5-cargo in the same importomer. This always leaves at least one PEX5 associated with each importomer.
) can only occur when coupled to the cargo translocation of a distinct PEX5-cargo in the same importomer. This always leaves at least one PEX5 associated with each importomer.
 . Different numbers of binding sites per importomer are shown from
. Different numbers of binding sites per importomer are shown from  (orange triangles) to
(orange triangles) to  (green diamonds), as shown in the legend; the legend also applies to (B), (C), and (D). The dashed black line is the measured cytosolic PEX5 concentration of
(green diamonds), as shown in the legend; the legend also applies to (B), (C), and (D). The dashed black line is the measured cytosolic PEX5 concentration of  . This is consistent with
. This is consistent with  when
when  . (B) Peroxisomal PEX5 fraction vs.
. (B) Peroxisomal PEX5 fraction vs.  . (C) Fraction of peroxisomal PEX5 that is ubiquitinated vs. PEX5 cargo addition rate,
. (C) Fraction of peroxisomal PEX5 that is ubiquitinated vs. PEX5 cargo addition rate,  . (D) Ubiquitin per peroxisome vs.
. (D) Ubiquitin per peroxisome vs.  . A characteristic increase of ubiquitination with
. A characteristic increase of ubiquitination with  is seen that is largely independent of the number of binding sites
is seen that is largely independent of the number of binding sites  . Vertical bars represent the standard deviation of observed values; error bars are smaller than point sizes.
. Vertical bars represent the standard deviation of observed values; error bars are smaller than point sizes.
 . The dashed black line is the measured cytosolic PEX5 concentration of
. The dashed black line is the measured cytosolic PEX5 concentration of  . Inset shows the fraction of importomers that are fully occupied by PEX5 vs. PEX5 cargo addition rate, with five PEX5 sites per importomer and cooperative coupling. (B) peroxisomal PEX5 fraction vs.
. Inset shows the fraction of importomers that are fully occupied by PEX5 vs. PEX5 cargo addition rate, with five PEX5 sites per importomer and cooperative coupling. (B) peroxisomal PEX5 fraction vs.  for cooperatively coupled cargo translocation. (C) Fraction of peroxisomal PEX5 that is ubiquitinated vs.
for cooperatively coupled cargo translocation. (C) Fraction of peroxisomal PEX5 that is ubiquitinated vs.  . (D) ubiquitin per peroxisome vs.
. (D) ubiquitin per peroxisome vs.  . A characteristic decrease of ubiquitination with
. A characteristic decrease of ubiquitination with  is seen that is largely independent of the number of binding sites
is seen that is largely independent of the number of binding sites  . Different number of binding sites per importomer are shown from
. Different number of binding sites per importomer are shown from  (red circles) to
(red circles) to  (green diamonds), as shown in the legend in (B). Cooperative coupling cannot function with
(green diamonds), as shown in the legend in (B). Cooperative coupling cannot function with  , so that is not shown. Subsequent figures use
, so that is not shown. Subsequent figures use  (blue squares). Note that the vertical scale of ubiquitin per peroxisome in (D) is much larger than in Fig. 3.
(blue squares). Note that the vertical scale of ubiquitin per peroxisome in (D) is much larger than in Fig. 3.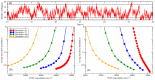
 , with the default number of peroxisomes (
, with the default number of peroxisomes ( ) and importomers per peroxisome (
) and importomers per peroxisome ( ). The characteristic timescale for fluctuations in the ubiquitination level is several seconds. Two possible threshold values are illustrated with dashed lines. (B) The average interval of time spent below a given threshold vs.
). The characteristic timescale for fluctuations in the ubiquitination level is several seconds. Two possible threshold values are illustrated with dashed lines. (B) The average interval of time spent below a given threshold vs.  for thresholds as indicated by the legend, which also applies to (C). (C) The average interval of time spent above a given threshold vs.
for thresholds as indicated by the legend, which also applies to (C). (C) The average interval of time spent above a given threshold vs.  .
.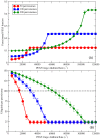
 , as indicated by legend in (A)) when the other parameters are kept constant (with
, as indicated by legend in (A)) when the other parameters are kept constant (with  sites per importomer). (A) Peroxisomal PEX5 fraction vs.
sites per importomer). (A) Peroxisomal PEX5 fraction vs.  for cooperatively coupled cargo translocation. (B) Ubiquitin per peroxisome vs.
for cooperatively coupled cargo translocation. (B) Ubiquitin per peroxisome vs.  . Horizontal black dashed line represents a possible ubiquitin threshold for peroxisome degradation.
. Horizontal black dashed line represents a possible ubiquitin threshold for peroxisome degradation.
 ,
,  , and
, and  we vary the number of export complexes
we vary the number of export complexes  , which directly scales the PEX5 export rate,
, which directly scales the PEX5 export rate,  . (A) Peroxisomal PEX5 fraction vs. stoichiometry of export complexes to importomers (
. (A) Peroxisomal PEX5 fraction vs. stoichiometry of export complexes to importomers ( ). As shown in the legend, we consider different fixed rates of cargo addition,
). As shown in the legend, we consider different fixed rates of cargo addition,  ; this legend also applies to (B). (B) Ubiquitin per peroxisome vs.
; this legend also applies to (B). (B) Ubiquitin per peroxisome vs.  , for the same set of
, for the same set of  .
.Similar articles
-
Export-deficient monoubiquitinated PEX5 triggers peroxisome removal in SV40 large T antigen-transformed mouse embryonic fibroblasts.Autophagy. 2015;11(8):1326-40. doi: 10.1080/15548627.2015.1061846. Autophagy. 2015. PMID: 26086376 Free PMC article.
-
A cargo-centered perspective on the PEX5 receptor-mediated peroxisomal protein import pathway.J Biol Chem. 2013 Oct 4;288(40):29151-9. doi: 10.1074/jbc.M113.487140. Epub 2013 Aug 20. J Biol Chem. 2013. PMID: 23963456 Free PMC article.
-
PEX5 translocation into and out of peroxisomes drives matrix protein import.Mol Cell. 2022 Sep 1;82(17):3209-3225.e7. doi: 10.1016/j.molcel.2022.07.004. Epub 2022 Aug 4. Mol Cell. 2022. PMID: 35931083 Free PMC article.
-
Role of PEX5 ubiquitination in maintaining peroxisome dynamics and homeostasis.Cell Cycle. 2017;16(21):2037-2045. doi: 10.1080/15384101.2017.1376149. Epub 2017 Sep 21. Cell Cycle. 2017. PMID: 28933989 Free PMC article. Review.
-
Peroxisomal matrix protein receptor ubiquitination and recycling.Biochim Biophys Acta. 2006 Dec;1763(12):1620-8. doi: 10.1016/j.bbamcr.2006.08.046. Epub 2006 Sep 3. Biochim Biophys Acta. 2006. PMID: 17028012 Review.
Cited by
-
Export-deficient monoubiquitinated PEX5 triggers peroxisome removal in SV40 large T antigen-transformed mouse embryonic fibroblasts.Autophagy. 2015;11(8):1326-40. doi: 10.1080/15548627.2015.1061846. Autophagy. 2015. PMID: 26086376 Free PMC article.
-
ATM functions at the peroxisome to induce pexophagy in response to ROS.Nat Cell Biol. 2015 Oct;17(10):1259-1269. doi: 10.1038/ncb3230. Epub 2015 Sep 7. Nat Cell Biol. 2015. PMID: 26344566 Free PMC article.
-
The cytosolic domain of Pex22p stimulates the Pex4p-dependent ubiquitination of the PTS1-receptor.PLoS One. 2014 Aug 27;9(8):e105894. doi: 10.1371/journal.pone.0105894. eCollection 2014. PLoS One. 2014. PMID: 25162638 Free PMC article.
-
Covalent Label Transfer between Peroxisomal Importomer Components Reveals Export-driven Import Interactions.J Biol Chem. 2016 Jan 29;291(5):2460-8. doi: 10.1074/jbc.M115.686501. Epub 2015 Nov 13. J Biol Chem. 2016. PMID: 26567336 Free PMC article.
-
Cysteine-specific ubiquitination protects the peroxisomal import receptor Pex5p against proteasomal degradation.Biosci Rep. 2015 May 14;35(3):e00215. doi: 10.1042/BSR20150103. Biosci Rep. 2015. PMID: 26182377 Free PMC article.
References
-
- Wanders RJA, Waterham HR (2006) Biochemistry of mammalian peroxisomes revisited. Annu Rev Biochem 75: 295–332 - PubMed
-
- Waterham HR, Ebberink MS (2012) Genetics and molecular basis of human peroxisome biogenesis disorders. Biochim Biophys Acta 1822: 1430–1441 - PubMed
-
- Platta HW, Erdmann R (2007) Peroxisomal dynamics. Trends Cell Biol 17: 474–484 - PubMed
-
- Hess R, Staubli W, Riess W (1965) Nature of the hepatomegalic effect produced by ethylchlorophenoxy-isobutyrate in the rat. Nature 208: 856–858 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

