PredictSNP: robust and accurate consensus classifier for prediction of disease-related mutations
- PMID: 24453961
- PMCID: PMC3894168
- DOI: 10.1371/journal.pcbi.1003440
PredictSNP: robust and accurate consensus classifier for prediction of disease-related mutations
Abstract
Single nucleotide variants represent a prevalent form of genetic variation. Mutations in the coding regions are frequently associated with the development of various genetic diseases. Computational tools for the prediction of the effects of mutations on protein function are very important for analysis of single nucleotide variants and their prioritization for experimental characterization. Many computational tools are already widely employed for this purpose. Unfortunately, their comparison and further improvement is hindered by large overlaps between the training datasets and benchmark datasets, which lead to biased and overly optimistic reported performances. In this study, we have constructed three independent datasets by removing all duplicities, inconsistencies and mutations previously used in the training of evaluated tools. The benchmark dataset containing over 43,000 mutations was employed for the unbiased evaluation of eight established prediction tools: MAPP, nsSNPAnalyzer, PANTHER, PhD-SNP, PolyPhen-1, PolyPhen-2, SIFT and SNAP. The six best performing tools were combined into a consensus classifier PredictSNP, resulting into significantly improved prediction performance, and at the same time returned results for all mutations, confirming that consensus prediction represents an accurate and robust alternative to the predictions delivered by individual tools. A user-friendly web interface enables easy access to all eight prediction tools, the consensus classifier PredictSNP and annotations from the Protein Mutant Database and the UniProt database. The web server and the datasets are freely available to the academic community at http://loschmidt.chemi.muni.cz/predictsnp.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
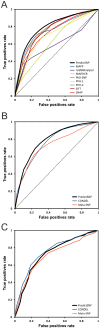
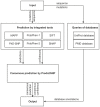

References
-
- Collins FS, Brooks LD, Chakravarti A (1998) A DNA polymorphism discovery resource for research on human genetic variation. Genome Res 8: 1229–1231 - PubMed
-
- Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, et al. (2010) A map of human genome variation from population-scale sequencing. Nature 467: 1061–1073doi:10.1038/nature09534 - DOI - PMC - PubMed
-
- Collins FS, Guyer MS, Charkravarti A (1997) Variations on a theme: cataloging human DNA sequence variation. Science 278: 1580–1581 - PubMed
-
- Risch N, Merikangas K (1996) The future of genetic studies of complex human diseases. Science 273: 1516–1517 - PubMed
-
- Studer RA, Dessailly BH, Orengo CA (2013) Residue mutations and their impact on protein structure and function: detecting beneficial and pathogenic changes. Biochem J 449: 581–594doi:10.1042/BJ20121221 - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

