Gammaherpesviral gene expression and virion composition are broadly controlled by accelerated mRNA degradation
- PMID: 24453974
- PMCID: PMC3894220
- DOI: 10.1371/journal.ppat.1003882
Gammaherpesviral gene expression and virion composition are broadly controlled by accelerated mRNA degradation
Abstract
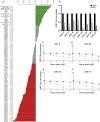
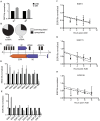
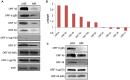
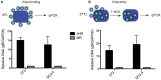
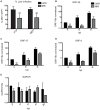
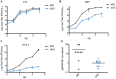
Lytic gammaherpesvirus infection restricts host gene expression by promoting widespread degradation of cytoplasmic mRNA through the activity of the viral endonuclease SOX. Though generally assumed to be selective for cellular transcripts, the extent to which SOX impacts viral mRNA stability has remained unknown. We addressed this issue using the model murine gammaherpesvirus MHV68 and, unexpectedly, found that all stages of viral gene expression are controlled through mRNA degradation. Using both comprehensive RNA expression profiling and half-life studies we reveal that the levels of the majority of viral mRNAs but not noncoding RNAs are tempered by MHV68 SOX (muSOX) activity. The targeting of viral mRNA by muSOX is functionally significant, as it impacts intracellular viral protein abundance and progeny virion composition. In the absence of muSOX-imposed gene expression control the viral particles display increased cell surface binding and entry as well as enhanced immediate early gene expression. These phenotypes culminate in a viral replication defect in multiple cell types as well as in vivo, highlighting the importance of maintaining the appropriate balance of viral RNA during gammaherpesviral infection. This is the first example of a virus that fails to broadly discriminate between cellular and viral transcripts during host shutoff and instead uses the targeting of viral messages to fine-tune overall gene expression.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

