A comprehensive tRNA deletion library unravels the genetic architecture of the tRNA pool
- PMID: 24453985
- PMCID: PMC3894157
- DOI: 10.1371/journal.pgen.1004084
A comprehensive tRNA deletion library unravels the genetic architecture of the tRNA pool
Abstract
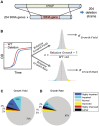
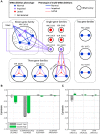
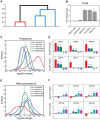
Deciphering the architecture of the tRNA pool is a prime challenge in translation research, as tRNAs govern the efficiency and accuracy of the process. Towards this challenge, we created a systematic tRNA deletion library in Saccharomyces cerevisiae, aimed at dissecting the specific contribution of each tRNA gene to the tRNA pool and to the cell's fitness. By harnessing this resource, we observed that the majority of tRNA deletions show no appreciable phenotype in rich medium, yet under more challenging conditions, additional phenotypes were observed. Robustness to tRNA gene deletion was often facilitated through extensive backup compensation within and between tRNA families. Interestingly, we found that within tRNA families, genes carrying identical anti-codons can contribute differently to the cellular fitness, suggesting the importance of the genomic surrounding to tRNA expression. Characterization of the transcriptome response to deletions of tRNA genes exposed two disparate patterns: in single-copy families, deletions elicited a stress response; in deletions of genes from multi-copy families, expression of the translation machinery increased. Our results uncover the complex architecture of the tRNA pool and pave the way towards complete understanding of their role in cell physiology.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Kozak M (2005) Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene 361: 13–37 doi:10.1016/j.gene.2005.06.037 - DOI - PubMed
-
- Jackson RJ, Hellen CUT, Pestova TV (2010) The mechanism of eukaryotic translation initiation and principles of its regulation. Nature reviews Molecular cell biology 11: 113–127 doi:10.1038/nrm2838 - DOI - PMC - PubMed
-
- Varenne S, Buc J, Lloubes R, Lazdunski C (1984) Translation is a non-uniform process. Effect of tRNA availability on the rate of elongation of nascent polypeptide chains. Journal of molecular biology 180: 549–576. - PubMed
-
- Kudla G, Murray AW, Tollervey D, Plotkin JB (2009) Coding-sequence determinants of gene expression in Escherichia coli. Science (New York, NY) 324: 255–258 doi:10.1126/science.1170160 - DOI - PMC - PubMed
-
- Stoletzki N, Eyre-Walker A (2007) Synonymous codon usage in Escherichia coli: selection for translational accuracy. Molecular biology and evolution 24: 374–381 doi:10.1093/molbev/msl166 - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

