Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells
- PMID: 24454679
- PMCID: PMC3894192
- DOI: 10.1371/journal.pone.0078644
Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells
Abstract
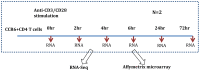
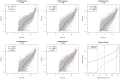
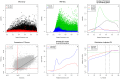
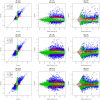
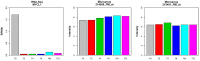
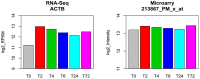
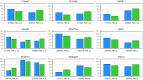
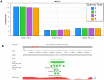
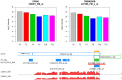
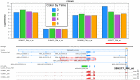
To demonstrate the benefits of RNA-Seq over microarray in transcriptome profiling, both RNA-Seq and microarray analyses were performed on RNA samples from a human T cell activation experiment. In contrast to other reports, our analyses focused on the difference, rather than similarity, between RNA-Seq and microarray technologies in transcriptome profiling. A comparison of data sets derived from RNA-Seq and Affymetrix platforms using the same set of samples showed a high correlation between gene expression profiles generated by the two platforms. However, it also demonstrated that RNA-Seq was superior in detecting low abundance transcripts, differentiating biologically critical isoforms, and allowing the identification of genetic variants. RNA-Seq also demonstrated a broader dynamic range than microarray, which allowed for the detection of more differentially expressed genes with higher fold-change. Analysis of the two datasets also showed the benefit derived from avoidance of technical issues inherent to microarray probe performance such as cross-hybridization, non-specific hybridization and limited detection range of individual probes. Because RNA-Seq does not rely on a pre-designed complement sequence detection probe, it is devoid of issues associated with probe redundancy and annotation, which simplified interpretation of the data. Despite the superior benefits of RNA-Seq, microarrays are still the more common choice of researchers when conducting transcriptional profiling experiments. This is likely because RNA-Seq sequencing technology is new to most researchers, more expensive than microarray, data storage is more challenging and analysis is more complex. We expect that once these barriers are overcome, the RNA-Seq platform will become the predominant tool for transcriptome analysis.
Conflict of interest statement
Figures


References
-
- Baldi P, Hatfield W (2002) DNA Microarray and Gene Expression. From experiments to data analysis and modeling. Cambridge University Press.
-
- Speed T, editor (2003) Statistical Analysis of Gene Expression Microarray Data. Chapman & Hall/CRC.
-
- Kerr G, Ruskin HJ, Crane M, Doolan P (2008) Techniques for clustering gene expression data. Computers in Biology and Medicine 38: 283–293. - PubMed
-
- Passador-Gurgel G, Hsieh W, Hunt P, Deighton N, Gibson G (2007) Quantitative trait transcripts for nicotine resistance in Drosophila melanogaster. Nat Genet 39: 264–268. - PubMed
-
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mam-malian transcriptomes by RNA-Seq. Nat Methods 5: 621–628. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- SRA/SRP026389
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

