Comparative structural and functional analysis of orthomyxovirus polymerase cap-snatching domains
- PMID: 24454773
- PMCID: PMC3893164
- DOI: 10.1371/journal.pone.0084973
Comparative structural and functional analysis of orthomyxovirus polymerase cap-snatching domains
Abstract
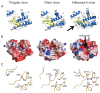
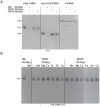
Orthomyxovirus Influenza A virus (IAV) heterotrimeric polymerase performs transcription of viral mRNAs by cap-snatching, which involves generation of capped primers by host pre-mRNA binding via the PB2 subunit cap-binding site and cleavage 10-13 nucleotides from the 5' cap by the PA subunit endonuclease. Thogotoviruses, tick-borne orthomyxoviruses that includes Thogoto (THOV), Dhori (DHOV) and Jos (JOSV) viruses, are thought to perform cap-snatching by cleaving directly after the cap and thus have no heterogeneous, host-derived sequences at the 5' extremity of their mRNAs. Based on recent work identifying the cap-binding and endonuclease domains in IAV polymerase, we determined the crystal structures of two THOV PB2 domains, the putative cap-binding and the so-called '627-domain', and the structures of the putative endonuclease domains (PA-Nter) of THOV and DHOV. Despite low sequence similarity, corresponding domains have the same fold confirming the overall architectural similarity of orthomyxovirus polymerases. However the putative Thogotovirus cap-snatching domains in PA and PB2 have non-conservative substitutions of key active site residues. Biochemical analysis confirms that, unlike the IAV domains, the THOV and DHOV PA-Nter domains do not bind divalent cations and have no endonuclease activity and the THOV central PB2 domain does not bind cap analogues. On the other hand, sequence analysis suggests that other, non-influenza, orthomyxoviruses, such as salmon anemia virus (isavirus) and Quaranfil virus likely conserve active cap-snatching domains correlating with the reported occurrence of heterogeneous, host-derived sequences at the 5' end of the mRNAs of these viruses. These results highlight the unusual nature of transcription initiation by Thogotoviruses.
Conflict of interest statement
Figures




References
-
- Kawaoka Y, Palese P (2005) Family Orthomyxoviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball L, editors. Virus Taxonomy: Eighth Report of the International Committee on Taxonomy of Viruses,. San Diego: Elsevier Academic Press. pp. 681–693.
-
- Haig DA, Woodall JP, Danskin D (1965) Thogoto Virus: A Hitherto Underscribed Agent Isolated from Ticks in Kenya. Journal of general microbiology 38: 389–394. - PubMed
-
- Anderson CR, Casals J (1973) Dhori virus, a new agent isolated from Hyalomma dromedarii in India. The Indian journal of medical research 61: 1416–1420. - PubMed
-
- Moore DL, Causey OR, Carey DE, Reddy S, Cooke AR, et al. (1975) Arthropod-borne viral infections of man in Nigeria, 1964–1970. Ann Trop Med Parasitol 69: 49–64. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

