Transposon defense by endo-siRNAs, piRNAs and somatic pilRNAs in Drosophila: contributions of Loqs-PD and R2D2
- PMID: 24454776
- PMCID: PMC3890300
- DOI: 10.1371/journal.pone.0084994
Transposon defense by endo-siRNAs, piRNAs and somatic pilRNAs in Drosophila: contributions of Loqs-PD and R2D2
Abstract
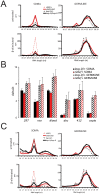
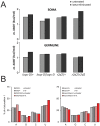
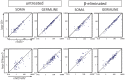
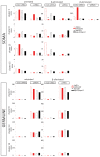
Transposable elements are a serious threat for genome integrity and their control via small RNA mediated silencing pathways is an ancient strategy. The fruit fly Drosophila melanogaster has two silencing activities that target transposons: endogenous siRNAs (esiRNAs or endo-siRNAs) and Piwi-interacting small RNAs (piRNAs). The biogenesis of endo-siRNAs involves the Dicer-2 co-factors Loqs-PD, which acts predominantly during processing of dsRNA by Dcr-2, and R2D2, which primarily helps to direct siRNAs into the RNA interference effector Ago2. Nonetheless, loss of either protein is not sufficient to produce a phenotype comparable with a dcr-2 mutation. We provide further deep sequencing evidence supporting the notion that R2D2 and Loqs-PD have partially overlapping function. Certain transposons display a preference for either dsRBD-protein during production or loading; this appeared to correlate neither with overall abundance, classification of the transposon or a specific site of genomic origin. The endo-siRNA biogenesis pathway in germline operates according to the same principles as the existing model for the soma, and its impairment does not significantly affect piRNAs. Expanding the analysis, we confirmed the occurrence of somatic piRNA-like RNAs (pilRNAs) that show a ping-pong signature. We detected expression of the Piwi-family protein mRNAs only barely above background, indicating that the somatic pilRNAs may arise from a small sub-population of somatic cells that express a functional piRNA pathway.
Conflict of interest statement
Figures
Similar articles
-
Processing of Drosophila endo-siRNAs depends on a specific Loquacious isoform.RNA. 2009 Oct;15(10):1886-95. doi: 10.1261/rna.1611309. Epub 2009 Jul 27. RNA. 2009. PMID: 19635780 Free PMC article.
-
Loqs-PD and R2D2 define independent pathways for RISC generation in Drosophila.Nucleic Acids Res. 2011 May;39(9):3836-51. doi: 10.1093/nar/gkq1324. Epub 2011 Jan 17. Nucleic Acids Res. 2011. PMID: 21245036 Free PMC article.
-
Maternally inherited piRNAs direct transient heterochromatin formation at active transposons during early Drosophila embryogenesis.Elife. 2021 Jul 8;10:e68573. doi: 10.7554/eLife.68573. Elife. 2021. PMID: 34236313 Free PMC article.
-
The piRNA pathway in Drosophila ovarian germ and somatic cells.Proc Jpn Acad Ser B Phys Biol Sci. 2020;96(1):32-42. doi: 10.2183/pjab.96.003. Proc Jpn Acad Ser B Phys Biol Sci. 2020. PMID: 31932527 Free PMC article. Review.
-
Untangling the web: the diverse functions of the PIWI/piRNA pathway.Mol Reprod Dev. 2013 Aug;80(8):632-64. doi: 10.1002/mrd.22195. Epub 2013 Jun 27. Mol Reprod Dev. 2013. PMID: 23712694 Free PMC article. Review.
Cited by
-
Drosophila dicer-2 cleavage is mediated by helicase- and dsRNA termini-dependent states that are modulated by Loquacious-PD.Mol Cell. 2015 May 7;58(3):406-17. doi: 10.1016/j.molcel.2015.03.012. Epub 2015 Apr 16. Mol Cell. 2015. PMID: 25891075 Free PMC article.
-
Changes of Osvaldo expression patterns in germline of male hybrids between the species Drosophila buzzatii and Drosophila koepferae.Mol Genet Genomics. 2015 Aug;290(4):1471-83. doi: 10.1007/s00438-015-1012-z. Epub 2015 Feb 25. Mol Genet Genomics. 2015. PMID: 25711309
-
Viral infection impacts transposable element transcript amounts in Drosophila.Proc Natl Acad Sci U S A. 2020 Jun 2;117(22):12249-12257. doi: 10.1073/pnas.2006106117. Epub 2020 May 20. Proc Natl Acad Sci U S A. 2020. PMID: 32434916 Free PMC article.
-
Expression of the Retrotransposon Helena Reveals a Complex Pattern of TE Deregulation in Drosophila Hybrids.PLoS One. 2016 Jan 26;11(1):e0147903. doi: 10.1371/journal.pone.0147903. eCollection 2016. PLoS One. 2016. PMID: 26812285 Free PMC article.
-
Specific PIWI-Interacting RNAs and Related Small Noncoding RNAs Are Associated With Ovarian Aging in Ames Dwarf (df/df) Mice.J Gerontol A Biol Sci Med Sci. 2021 Aug 13;76(9):1561-1570. doi: 10.1093/gerona/glab113. J Gerontol A Biol Sci Med Sci. 2021. PMID: 34387333 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

