Diversity of Prdm9 zinc finger array in wild mice unravels new facets of the evolutionary turnover of this coding minisatellite
- PMID: 24454780
- PMCID: PMC3890296
- DOI: 10.1371/journal.pone.0085021
Diversity of Prdm9 zinc finger array in wild mice unravels new facets of the evolutionary turnover of this coding minisatellite
Abstract
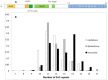
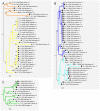
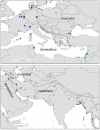
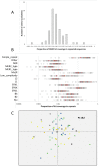
In humans and mice, meiotic recombination events cluster into narrow hotspots whose genomic positions are defined by the PRDM9 protein via its DNA binding domain constituted of an array of zinc fingers (ZnFs). High polymorphism and rapid divergence of the Prdm9 gene ZnF domain appear to involve positive selection at DNA-recognition amino-acid positions, but the nature of the underlying evolutionary pressures remains a puzzle. Here we explore the variability of the Prdm9 ZnF array in wild mice, and uncovered a high allelic diversity of both ZnF copy number and identity with the caracterization of 113 alleles. We analyze features of the diversity of ZnF identity which is mostly due to non-synonymous changes at codons -1, 3 and 6 of each ZnF, corresponding to amino-acids involved in DNA binding. Using methods adapted to the minisatellite structure of the ZnF array, we infer a phylogenetic tree of these alleles. We find the sister species Mus spicilegus and M. macedonicus as well as the three house mouse (Mus musculus) subspecies to be polyphyletic. However some sublineages have expanded independently in Mus musculus musculus and M. m. domesticus, the latter further showing phylogeographic substructure. Compared to random genomic regions and non-coding minisatellites, none of these patterns appears exceptional. In silico prediction of DNA binding sites for each allele, overlap of their alignments to the genome and relative coverage of the different families of interspersed repeated elements suggest a large diversity between PRDM9 variants with a potential for highly divergent distributions of recombination events in the genome with little correlation to evolutionary distance. By compiling PRDM9 ZnF protein sequences in Primates, Muridae and Equids, we find different diversity patterns among the three amino-acids most critical for the DNA-recognition function, suggesting different diversification timescales.
Conflict of interest statement
Figures


Similar articles
-
Rapid Evolution of the Fine-scale Recombination Landscape in Wild House Mouse (Mus musculus) Populations.Mol Biol Evol. 2023 Jan 4;40(1):msac267. doi: 10.1093/molbev/msac267. Mol Biol Evol. 2023. PMID: 36508360 Free PMC article.
-
Prdm9 polymorphism unveils mouse evolutionary tracks.DNA Res. 2014 Jun;21(3):315-26. doi: 10.1093/dnares/dst059. Epub 2014 Jan 20. DNA Res. 2014. PMID: 24449848 Free PMC article.
-
The long zinc finger domain of PRDM9 forms a highly stable and long-lived complex with its DNA recognition sequence.Chromosome Res. 2017 Jun;25(2):155-172. doi: 10.1007/s10577-017-9552-1. Epub 2017 Feb 2. Chromosome Res. 2017. PMID: 28155083 Free PMC article.
-
The consequences of sequence erosion in the evolution of recombination hotspots.Philos Trans R Soc Lond B Biol Sci. 2017 Dec 19;372(1736):20160462. doi: 10.1098/rstb.2016.0462. Philos Trans R Soc Lond B Biol Sci. 2017. PMID: 29109225 Free PMC article. Review.
-
The case of the fickle fingers: how the PRDM9 zinc finger protein specifies meiotic recombination hotspots in humans.PLoS Biol. 2011 Dec;9(12):e1001211. doi: 10.1371/journal.pbio.1001211. Epub 2011 Dec 6. PLoS Biol. 2011. PMID: 22162947 Free PMC article. Review.
Cited by
-
Meiotic chromosome dynamics and double strand break formation in reptiles.Front Cell Dev Biol. 2022 Oct 12;10:1009776. doi: 10.3389/fcell.2022.1009776. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36313577 Free PMC article.
-
Rapid Evolution of the Fine-scale Recombination Landscape in Wild House Mouse (Mus musculus) Populations.Mol Biol Evol. 2023 Jan 4;40(1):msac267. doi: 10.1093/molbev/msac267. Mol Biol Evol. 2023. PMID: 36508360 Free PMC article.
-
PR Domain-containing Protein 7 (PRDM7) Is a Histone 3 Lysine 4 Trimethyltransferase.J Biol Chem. 2016 Jun 24;291(26):13509-19. doi: 10.1074/jbc.M116.721472. Epub 2016 Apr 29. J Biol Chem. 2016. PMID: 27129774 Free PMC article.
-
PRDM9 drives evolutionary erosion of hotspots in Mus musculus through haplotype-specific initiation of meiotic recombination.PLoS Genet. 2015 Jan 8;11(1):e1004916. doi: 10.1371/journal.pgen.1004916. eCollection 2015 Jan. PLoS Genet. 2015. PMID: 25568937 Free PMC article.
-
Evolution of the recombination regulator PRDM9 in minke whales.BMC Genomics. 2022 Mar 16;23(1):212. doi: 10.1186/s12864-022-08305-1. BMC Genomics. 2022. PMID: 35296233 Free PMC article.
References
-
- Handel MA, Schimenti JC (2010) Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat Rev Genet 11: 124–136. - PubMed
-
- McKee BD (2009) Homolog pairing and segregation in Drosophila meiosis. Genome Dyn 5: 56–68. - PubMed
-
- Otto SP, Lenormand T (2002) Resolving the paradox of sex and recombination. Nat Rev Genet 3: 252–261. - PubMed
-
- Coop G, Przeworski M (2007) An evolutionary view of human recombination. Nat Rev Genet 8: 23–34. - PubMed
-
- Baudat F, de Massy B (2007) Regulating double-stranded DNA break repair towards crossover or non-crossover during mammalian meiosis. Chromosome Res 15: 565–577. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

