A simple lattice model that captures protein folding, aggregation and amyloid formation
- PMID: 24454816
- PMCID: PMC3893179
- DOI: 10.1371/journal.pone.0085185
A simple lattice model that captures protein folding, aggregation and amyloid formation
Abstract
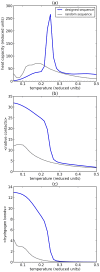
The ability of many proteins to convert from their functional soluble state to amyloid fibrils can be attributed to inter-molecular beta strand formation. Such amyloid formation is associated with neurodegenerative disorders like Alzheimer's and Parkinson's. Molecular modelling can play a key role in providing insight into the factors that make proteins prone to fibril formation. However, fully atomistic models are computationally too expensive to capture the length and time scales associated with fibril formation. As the ability to form fibrils is the rule rather than the exception, much insight can be gained from the study of coarse-grained models that capture the key generic features associated with amyloid formation. Here we present a simple lattice model that can capture both protein folding and beta strand formation. Unlike standard lattice models, this model explicitly incorporates the formation of hydrogen bonds and the directionality of side chains. The simplicity of our model makes it computationally feasible to investigate the interplay between folding, amorphous aggregation and fibril formation, and maintains the capability of classic lattice models to simulate protein folding with high specificity. In our model, the folded proteins contain structures that resemble naturally occurring beta-sheets, with alternating polar and hydrophobic amino acids. Moreover, fibrils with intermolecular cross-beta strand conformations can be formed spontaneously out of multiple short hydrophobic peptide sequences. Both the formation of hydrogen bonds in folded structures and in fibrils is strongly dependent on the amino acid sequence, indicating that hydrogen-bonding interactions alone are not strong enough to initiate the formation of beta sheets. This result agrees with experimental observations that beta sheet and amyloid formation is strongly sequence dependent, with hydrophobic sequences being more prone to form such structures. Our model should open the way to a systematic study of the interplay between the factors that lead to amyloid formation.
Conflict of interest statement
Figures
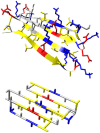


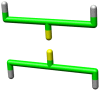
 ), only fibrils that could form a strong hydrophobic core (TFTFTFT) are stable, in this case the hydrophobic residues would point inwards, as shown in Figure 3.
), only fibrils that could form a strong hydrophobic core (TFTFTFT) are stable, in this case the hydrophobic residues would point inwards, as shown in Figure 3.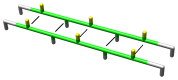

Similar articles
-
Role of water in protein aggregation and amyloid polymorphism.Acc Chem Res. 2012 Jan 17;45(1):83-92. doi: 10.1021/ar2000869. Epub 2011 Jul 15. Acc Chem Res. 2012. PMID: 21761818 Free PMC article.
-
Molecular structures of amyloid and prion fibrils: consensus versus controversy.Acc Chem Res. 2013 Jul 16;46(7):1487-96. doi: 10.1021/ar300282r. Epub 2013 Jan 7. Acc Chem Res. 2013. PMID: 23294335 Free PMC article. Review.
-
Protein denaturation and aggregation: Cellular responses to denatured and aggregated proteins.Ann N Y Acad Sci. 2005 Dec;1066:181-221. doi: 10.1196/annals.1363.030. Ann N Y Acad Sci. 2005. PMID: 16533927 Review.
-
Self-folding and aggregation of amyloid nanofibrils.Nanoscale. 2011 Apr;3(4):1748-55. doi: 10.1039/c0nr00840k. Epub 2011 Feb 23. Nanoscale. 2011. PMID: 21347488
-
Elucidating the Structures of Amyloid Oligomers with Macrocyclic β-Hairpin Peptides: Insights into Alzheimer's Disease and Other Amyloid Diseases.Acc Chem Res. 2018 Mar 20;51(3):706-718. doi: 10.1021/acs.accounts.7b00554. Epub 2018 Mar 6. Acc Chem Res. 2018. PMID: 29508987 Free PMC article. Review.
Cited by
-
Multi-eGO: An in silico lens to look into protein aggregation kinetics at atomic resolution.Proc Natl Acad Sci U S A. 2022 Jun 28;119(26):e2203181119. doi: 10.1073/pnas.2203181119. Epub 2022 Jun 23. Proc Natl Acad Sci U S A. 2022. PMID: 35737839 Free PMC article.
-
Knotted proteins: A tangled tale of Structural Biology.Comput Struct Biotechnol J. 2015 Aug 19;13:459-68. doi: 10.1016/j.csbj.2015.08.003. eCollection 2015. Comput Struct Biotechnol J. 2015. PMID: 26380658 Free PMC article. Review.
-
Evaluation of FRET X for single-molecule protein fingerprinting.iScience. 2021 Oct 5;24(11):103239. doi: 10.1016/j.isci.2021.103239. eCollection 2021 Nov 19. iScience. 2021. PMID: 34729466 Free PMC article.
-
Unraveling the Nature of Hydrogen Bonds of "Proton Sponges" Based on Car-Parrinello and Metadynamics Approaches.Int J Mol Sci. 2023 Jan 12;24(2):1542. doi: 10.3390/ijms24021542. Int J Mol Sci. 2023. PMID: 36675059 Free PMC article.
-
Computational Models for the Study of Protein Aggregation.Methods Mol Biol. 2022;2340:51-78. doi: 10.1007/978-1-0716-1546-1_4. Methods Mol Biol. 2022. PMID: 35167070
References
-
- Dobson CM (2003) Protein folding and misfolding. Nature 426: 884–890. - PubMed
-
- Combe N, Frenkel D (2007) Simple off-lattice model to study the folding and aggregation of peptides. Mol Phys375385.
-
- Tsigelny IF, Bar-On P, Sharikov Y, Crews L, Hashimoto M, et al. (2007) Dynamics of alphasynuclein aggregation and inhibition of pore-like oligomer development by beta-synuclein. FEBS J 274: 1862–77. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

