Mutation and selection cause codon usage and bias in mitochondrial genomes of ribbon worms (Nemertea)
- PMID: 24454907
- PMCID: PMC3893253
- DOI: 10.1371/journal.pone.0085631
Mutation and selection cause codon usage and bias in mitochondrial genomes of ribbon worms (Nemertea)
Abstract
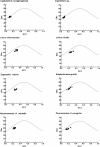
The phenomenon of codon usage bias is known to exist in many genomes and it is mainly determined by mutation and selection. To understand the patterns of codon usage in nemertean mitochondrial genomes, we use bioinformatic approaches to analyze the protein-coding sequences of eight nemertean species. Neutrality analysis did not find a significant correlation between GC12 and GC3. ENc-plot showed a few genes on or close to the expected curve, but the majority of points with low-ENc values are below it. ENc-plot suggested that mutational bias plays a major role in shaping codon usage. The Parity Rule 2 plot (PR2) analysis showed that GC and AT were not used proportionally and we propose that codons containing A or U at third position are used preferentially in nemertean species, regardless of whether corresponding tRNAs are encoded in the mitochondrial DNA. Context-dependent analysis indicated that the nucleotide at the second codon position slightly affects synonymous codon choices. These results suggested that mutational and selection forces are probably acting to codon usage bias in nemertean mitochondrial genomes.
Conflict of interest statement
Figures



Similar articles
-
Codon Usage Bias and Determining Forces in Taenia solium Genome.Korean J Parasitol. 2015 Dec;53(6):689-97. doi: 10.3347/kjp.2015.53.6.689. Epub 2015 Dec 31. Korean J Parasitol. 2015. PMID: 26797435 Free PMC article.
-
Analysis of codon usage bias of WRKY transcription factors in Helianthus annuus.BMC Genom Data. 2022 Jun 20;23(1):46. doi: 10.1186/s12863-022-01064-8. BMC Genom Data. 2022. PMID: 35725374 Free PMC article.
-
Analysis of codon usage pattern in Taenia saginata based on a transcriptome dataset.Parasit Vectors. 2014 Dec 2;7:527. doi: 10.1186/s13071-014-0527-1. Parasit Vectors. 2014. PMID: 25440955 Free PMC article.
-
Codon usage bias.Mol Biol Rep. 2022 Jan;49(1):539-565. doi: 10.1007/s11033-021-06749-4. Epub 2021 Nov 25. Mol Biol Rep. 2022. PMID: 34822069 Free PMC article. Review.
-
Codon usage and codon pair patterns in non-grass monocot genomes.Ann Bot. 2017 Nov 28;120(6):893-909. doi: 10.1093/aob/mcx112. Ann Bot. 2017. PMID: 29155926 Free PMC article. Review.
Cited by
-
Strong Selectional Forces Fine-Tune CpG Content in Genes Involved in Neurological Disorders as Revealed by Codon Usage Patterns.Front Neurosci. 2022 Jun 10;16:887929. doi: 10.3389/fnins.2022.887929. eCollection 2022. Front Neurosci. 2022. PMID: 35757545 Free PMC article.
-
Insight into Codon Utilization Pattern of Tumor Suppressor Gene EPB41L3 from Different Mammalian Species Indicates Dominant Role of Selection Force.Cancers (Basel). 2021 Jun 1;13(11):2739. doi: 10.3390/cancers13112739. Cancers (Basel). 2021. PMID: 34205890 Free PMC article.
-
Genome-Wide Analysis of Codon Usage Patterns of SARS-CoV-2 Virus Reveals Global Heterogeneity of COVID-19.Biomolecules. 2021 Jun 18;11(6):912. doi: 10.3390/biom11060912. Biomolecules. 2021. PMID: 34207362 Free PMC article.
-
Comprehensive Analysis of Codon Usage Bias in Human Papillomavirus Type 51.Pol J Microbiol. 2024 Oct 28;73(4):455-465. doi: 10.33073/pjm-2024-036. eCollection 2024 Dec 1. Pol J Microbiol. 2024. PMID: 39465910 Free PMC article.
-
The complete mitochondrial genome of Talpa martinorum (Mammalia: Talpidae), a mole species endemic to Thrace: genome content and phylogenetic considerations.Genetica. 2022 Oct;150(5):317-325. doi: 10.1007/s10709-022-00162-w. Epub 2022 Aug 27. Genetica. 2022. PMID: 36029420
References
-
- Ikemura T (1981) Correlation between the abundance of escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes. J Mol Biol 146: 1–21. - PubMed
-
- Ikemura T (1985) Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol 2: 13–34. - PubMed
-
- Akashi H, Eyre-Walker A (1998) Translational selection and molecular evolution. Curr Opin Genet Dev 8: 688–693. - PubMed
-
- Akashi H (2001) Gene expression and molecular evolution. Curr Opin Genet Dev 11: 660–666. - PubMed
-
- Duret L (2002) Evolution of synonymous codon usage in metazoans. Curr Opin Genet Dev 12: 640–649. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

