Imaging the directed transport of single engineered RNA transcripts in real-time using ratiometric bimolecular beacons
- PMID: 24454933
- PMCID: PMC3893274
- DOI: 10.1371/journal.pone.0085813
Imaging the directed transport of single engineered RNA transcripts in real-time using ratiometric bimolecular beacons
Abstract
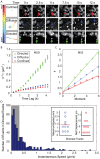
The relationship between RNA expression and cell function can often be difficult to decipher due to the presence of both temporal and sub-cellular processing of RNA. These intricacies of RNA regulation can often be overlooked when only acquiring global measurements of RNA expression. This has led to development of several tools that allow for the real-time imaging of individual engineered RNA transcripts in living cells. Here, we describe a new technique that utilizes an oligonucleotide-based probe, ratiometric bimolecular beacon (RBMB), to image RNA transcripts that were engineered to contain 96-tandem repeats of the RBMB target sequence in the 3'-untranslated region. Binding of RBMBs to the target RNA resulted in discrete bright fluorescent spots, representing individual transcripts, that could be imaged in real-time. Since RBMBs are a synthetic probe, the use of photostable, bright, and red-shifted fluorophores led to a high signal-to-background. RNA motion was readily characterized by both mean squared displacement and moment scaling spectrum analyses. These analyses revealed clear examples of directed, Brownian, and subdiffusive movements.
Conflict of interest statement
Figures
References
-
- Tyagi S (2009) Imaging intracellular RNA distribution and dynamics in living cells. Nat Methods 6: 331–338. - PubMed
-
- Cha BJ, Koppetsch BS, Theurkauf WE (2001) In vivo analysis of Drosophila bicoid mRNA localization reveals a novel microtubule-dependent axis specification pathway. Cell 106: 35–46. - PubMed
-
- Ishihama Y, Funatsu T (2009) Single molecule tracking of quantum dot-labeled mRNAs in a cell nucleus. Biochem Biophys Res Commun 381: 33–38. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

