Aptamers: problems, solutions and prospects
- PMID: 24455181
- PMCID: PMC3890987
Aptamers: problems, solutions and prospects
Abstract
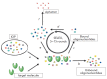
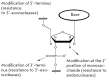
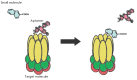
Aptamers are short single-stranded oligonucleotides that are capable of binding various molecules with high affinity and specificity. When the technology of aptamer selection was developed almost a quarter of a century ago, a suggestion was immediately put forward that it might be a revolutionary start into solving many problems associated with diagnostics and the therapy of diseases. However, multiple attempts to use aptamers in practice, although sometimes successful, have been generally much less efficient than had been expected initially. This review is mostly devoted not to the successful use of aptamers but to the problems impeding the widespread application of aptamers in diagnostics and therapy, as well as to approaches that could considerably expand the range of aptamer application.
Keywords: SELEX; aptamer; diagnostics; problems; therapeutics.
Figures



References
-
- Gilbert W., Nature. 1986;319:618.
LinkOut - more resources
Full Text Sources
Other Literature Sources
