Anatomy and transcript profiling of gynoecium development in female sterile Brassica napus mediated by one alien chromosome from Orychophragmus violaceus
- PMID: 24456102
- PMCID: PMC3930543
- DOI: 10.1186/1471-2164-15-61
Anatomy and transcript profiling of gynoecium development in female sterile Brassica napus mediated by one alien chromosome from Orychophragmus violaceus
Abstract
Background: The gynoecium is one of the most complex organs of angiosperms specialized for seed production and dispersal, but only several genes important for ovule or embryo sac development were identified by using female sterile mutants. The female sterility in oilseed rape (Brassica napus) was before found to be related with one alien chromosome from another crucifer Orychophragmus violaceus. Herein, the developmental anatomy and comparative transcript profiling (RNA-seq) for the female sterility were performed to reveal the genes and possible metabolic pathways behind the formation of the damaged gynoecium.
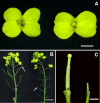
Results: The ovules in the female sterile Brassica napus with two copies of the alien chromosomes (S1) initiated only one short integument primordium which underwent no further development and the female gametophyte development was blocked after the tetrad stage but before megagametogenesis initiation. Using Brassica_ 95k_ unigene as the reference genome, a total of 28,065 and 27,653 unigenes were identified to be transcribed in S1 and donor B. napus (H3), respectively. Further comparison of the transcript abundance between S1 and H3 revealed that 4540 unigenes showed more than two fold expression differences. Gene ontology and pathway enrichment analysis of the Differentially Expressed Genes (DEGs) showed that a number of important genes and metabolism pathways were involved in the development of gynoecium, embryo sac, ovule, integuments as well as the interactions between pollen and pistil.
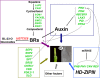
Conclusions: DEGs for the ovule development were detected to function in the metabolism pathways regulating brassinosteroid (BR) biosynthesis, adaxial/abaxial axis specification, auxin transport and signaling. A model was proposed to show the possible roles and interactions of these pathways for the sterile gynoecium development. The results provided new information for the molecular mechanisms behind the gynoecium development at early stage in B. napus.
Figures





References
-
- Kuusk S, Sohlberg JJ, Long JA, Fridborg I, Sundberg E. STY1 and STY2 promote the formation of apical tissues during Arabidopsis gynoecium development. Development. 2002;15(20):4707–4717. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

