Inhalable microorganisms in Beijing's PM2.5 and PM10 pollutants during a severe smog event
- PMID: 24456276
- PMCID: PMC3963435
- DOI: 10.1021/es4048472
Inhalable microorganisms in Beijing's PM2.5 and PM10 pollutants during a severe smog event
Abstract
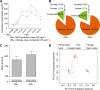
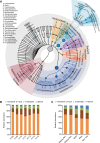
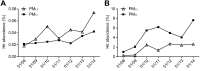
Particulate matter (PM) air pollution poses a formidable public health threat to the city of Beijing. Among the various hazards of PM pollutants, microorganisms in PM2.5 and PM10 are thought to be responsible for various allergies and for the spread of respiratory diseases. While the physical and chemical properties of PM pollutants have been extensively studied, much less is known about the inhalable microorganisms. Most existing data on airborne microbial communities using 16S or 18S rRNA gene sequencing to categorize bacteria or fungi into the family or genus levels do not provide information on their allergenic and pathogenic potentials. Here we employed metagenomic methods to analyze the microbial composition of Beijing's PM pollutants during a severe January smog event. We show that with sufficient sequencing depth, airborne microbes including bacteria, archaea, fungi, and dsDNA viruses can be identified at the species level. Our results suggested that the majority of the inhalable microorganisms were soil-associated and nonpathogenic to human. Nevertheless, the sequences of several respiratory microbial allergens and pathogens were identified and their relative abundance appeared to have increased with increased concentrations of PM pollution. Our findings may serve as an important reference for environmental scientists, health workers, and city planners.
Figures

References
-
- Cheng Z.; Jiang J.; Fajardo O.; Wang S.; Hao J. Characteristics and health impacts of particulate matter pollution in China (2001–2011). Atmos. Environ. 2013, 650186–194.
-
- Ouyang Y. China wakes up to the crisis of air pollution. Lancet Respir. Med. 2013, 1112. - PubMed
-
- Parrish D. D.; Zhu T. Climate change. Clean air for megacities. Science 2009, 3265953674–675. - PubMed
-
- Zhang Q.; He K.; Huo H. Policy: Cleaning China’s air. Nature 2012, 4847393161–162. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

