Perilous journey: a tour of the ubiquitin-proteasome system
- PMID: 24457024
- PMCID: PMC4037451
- DOI: 10.1016/j.tcb.2013.12.003
Perilous journey: a tour of the ubiquitin-proteasome system
Abstract
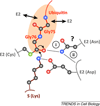
Eukaryotic cells are equipped to degrade proteins via the ubiquitin-proteasome system (UPS). Proteins become degraded upon their conjugation to chains of ubiquitin where they are then directed to the 26S proteasome, a macromolecular protease. The transfer of ubiquitin to proteins and their subsequent degradation are highly complex processes, and new research is beginning to uncover the molecular details of how ubiquitination and degradation take place in the cell. We review some of the new data providing insights into how these processes occur. Although distinct mechanisms are often observed, some common themes are emerging for how the UPS guides protein substrates through their final journey.
Keywords: E1-activating; E2-conjugating; E3-ligase; proteolysis; ubiquitin proteasome system.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures

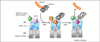
Similar articles
-
Ubiquitin-Proteasome Pathway and Muscle Atrophy.Adv Exp Med Biol. 2018;1088:235-248. doi: 10.1007/978-981-13-1435-3_10. Adv Exp Med Biol. 2018. PMID: 30390254 Review.
-
Ubiquitin proteasome system in immune regulation and therapeutics.Curr Opin Pharmacol. 2022 Dec;67:102310. doi: 10.1016/j.coph.2022.102310. Epub 2022 Oct 23. Curr Opin Pharmacol. 2022. PMID: 36288660 Free PMC article. Review.
-
A critical discussion on the relationship between E3 ubiquitin ligases, protein degradation, and skeletal muscle wasting: it's not that simple.Am J Physiol Cell Physiol. 2023 Dec 1;325(6):C1567-C1582. doi: 10.1152/ajpcell.00457.2023. Epub 2023 Nov 13. Am J Physiol Cell Physiol. 2023. PMID: 37955121 Free PMC article. Review.
-
Degradation of the stress-responsive enzyme formate dehydrogenase by the RING-type E3 ligase Keep on Going and the ubiquitin 26S proteasome system.Plant Mol Biol. 2018 Feb;96(3):265-278. doi: 10.1007/s11103-017-0691-8. Epub 2017 Dec 21. Plant Mol Biol. 2018. PMID: 29270890
-
A patent review of the ubiquitin ligase system: 2015-2018.Expert Opin Ther Pat. 2018 Dec;28(12):919-937. doi: 10.1080/13543776.2018.1549229. Epub 2018 Nov 23. Expert Opin Ther Pat. 2018. PMID: 30449221 Free PMC article. Review.
Cited by
-
The Stability and Expression Level of Bok Are Governed by Binding to Inositol 1,4,5-Trisphosphate Receptors.J Biol Chem. 2016 May 27;291(22):11820-8. doi: 10.1074/jbc.M115.711242. Epub 2016 Apr 6. J Biol Chem. 2016. PMID: 27053113 Free PMC article.
-
Visualizing K48 Ubiquitination during Presynaptic Formation By Ubiquitination-Induced Fluorescence Complementation (UiFC).Front Mol Neurosci. 2016 Jun 10;9:43. doi: 10.3389/fnmol.2016.00043. eCollection 2016. Front Mol Neurosci. 2016. PMID: 27375430 Free PMC article.
-
Trehalose Promotes Clearance of Proteotoxic Aggregation of Neurodegenerative Disease-Associated Aberrant Proteins.Mol Neurobiol. 2024 Jul;61(7):4055-4073. doi: 10.1007/s12035-023-03824-8. Epub 2023 Dec 7. Mol Neurobiol. 2024. PMID: 38057642
-
Proteolysis-targeting chimeras (PROTACs) in cancer therapy.Mol Cancer. 2022 Apr 11;21(1):99. doi: 10.1186/s12943-021-01434-3. Mol Cancer. 2022. PMID: 35410300 Free PMC article. Review.
-
Catalysis of non-canonical protein ubiquitylation by the ARIH1 ubiquitin ligase.Biochem J. 2023 Nov 29;480(22):1817-1831. doi: 10.1042/BCJ20230373. Biochem J. 2023. PMID: 37870100 Free PMC article.
References
-
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 2009;78:399–434. - PubMed
-
- Tokunaga F, et al. Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nat. Cell Biol. 2009;11:123–132. - PubMed
-
- Komander D, Rape M. The ubiquitin code. Annu. Rev. Biochem. 2012;81:203–229. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

