Genomic variability of monkeypox virus among humans, Democratic Republic of the Congo
- PMID: 24457084
- PMCID: PMC3901482
- DOI: 10.3201/eid2002.130118
Genomic variability of monkeypox virus among humans, Democratic Republic of the Congo
Abstract
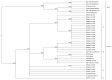
Monkeypox virus is a zoonotic virus endemic to Central Africa. Although active disease surveillance has assessed monkeypox disease prevalence and geographic range, information about virus diversity is lacking. We therefore assessed genome diversity of viruses in 60 samples obtained from humans with primary and secondary cases of infection from 2005 through 2007. We detected 4 distinct lineages and a deletion that resulted in gene loss in 10 (16.7%) samples and that seemed to correlate with human-to-human transmission (p = 0.0544). The data suggest a high frequency of spillover events from the pool of viruses in nonhuman animals, active selection through genomic destabilization and gene loss, and increased disease transmissibility and severity. The potential for accelerated adaptation to humans should be monitored through improved surveillance.
Keywords: Democratic Republic of the Congo; Monkeypox virus; emerging infectious disease; gene loss; genomic diversity; genomic reduction; viruses.
Figures

References
-
- Moss B. Poxviridae: The viruses and their replication. In: Knipe DM, Howley PM. Field's Virology, 4th ed. Philadelphia: Lippincott-Raven 2001. pp. 2849–83.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

