The fidelity of stochastic single-molecule super-resolution reconstructions critically depends upon robust background estimation
- PMID: 24458236
- PMCID: PMC3900998
- DOI: 10.1038/srep03854
The fidelity of stochastic single-molecule super-resolution reconstructions critically depends upon robust background estimation
Abstract
The quality of super resolution images obtained by stochastic single-molecule microscopy critically depends on image analysis algorithms. We find that the choice of background estimator is often the most important determinant of reconstruction quality. A variety of techniques have found use, but many have a very narrow range of applicability depending upon the characteristics of the raw data. Importantly, we observe that when using otherwise accurate algorithms, unaccounted background components can give rise to biases on scales defeating the purpose of super-resolution microscopy. We find that a temporal median filter in particular provides a simple yet effective solution to the problem of background estimation, which we demonstrate over a range of imaging modalities and different reconstruction methods.
Figures

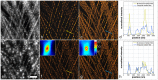

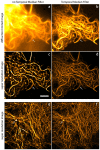

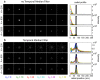
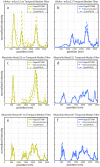
Comment in
-
Taming the image background beast.Nat Methods. 2014 Mar;11(3):228. doi: 10.1038/nmeth.2873. Nat Methods. 2014. PMID: 24724168 No abstract available.
References
-
- Ewers H. 4. Nano Resolution Optical Imaging Through Localization Microscopy. 1–20 (2012). 10.1016/b978-0-12-385872-6.00004-0.
-
- Wolter S., Löschberger A., Holm T. & Aufmkolk S. rapidSTORM: accurate, fast open-source software for localization microscopy. Nature (2012). - PubMed
-
- Henriques R. et al. QuickPALM: 3D real-time photoactivation nanoscopy image processing in ImageJ. Nature Methods 7, 339–340 (2010). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

