Small-molecule control of intracellular protein levels through modulation of the ubiquitin proteasome system
- PMID: 24459094
- PMCID: PMC4348030
- DOI: 10.1002/anie.201307761
Small-molecule control of intracellular protein levels through modulation of the ubiquitin proteasome system
Abstract
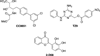
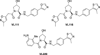
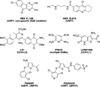
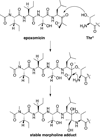
Traditionally, biological probes and drugs have targeted the activities of proteins (such as enzymes and receptors) that can be readily controlled by small molecules. The remaining majority of the proteome has been deemed "undruggable". By using small-molecule modulators of the ubiquitin proteasome, protein levels, rather than protein activity, can be targeted instead, thus increasing the number of druggable targets. Whereas targeting of the proteasome itself can lead to a global increase in protein levels, the targeting of other components of the UPS (e.g., the E3 ubiquitin ligases) can lead to an increase in protein levels in a more targeted fashion. Alternatively, multiple strategies for inducing protein degradation with small-molecule probes are emerging. With the ability to induce and inhibit the degradation of targeted proteins, small-molecule modulators of the UPS have the potential to significantly expand the druggable portion of the proteome beyond traditional targets, such as enzymes and receptors.
Keywords: drug design; inhibitors; proteasome; protein degradation; ubiquitin.
Copyright © 2014 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures















References
-
- Kravtsova-Ivantsiv Y, Sommer T, Ciechanover A. Angew. Chem. Int. Ed. 2013;52:192–198. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

