A complex of Cas proteins 5, 6, and 7 is required for the biogenesis and stability of clustered regularly interspaced short palindromic repeats (crispr)-derived rnas (crrnas) in Haloferax volcanii
- PMID: 24459147
- PMCID: PMC3945376
- DOI: 10.1074/jbc.M113.508184
A complex of Cas proteins 5, 6, and 7 is required for the biogenesis and stability of clustered regularly interspaced short palindromic repeats (crispr)-derived rnas (crrnas) in Haloferax volcanii
Abstract
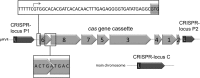
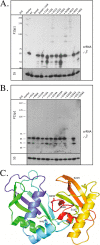
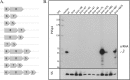
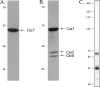
The clustered regularly interspaced short palindromic repeats/CRISPR-associated (CRISPR-Cas) system is a prokaryotic defense mechanism against foreign genetic elements. A plethora of CRISPR-Cas versions exist, with more than 40 different Cas protein families and several different molecular approaches to fight the invading DNA. One of the key players in the system is the CRISPR-derived RNA (crRNA), which directs the invader-degrading Cas protein complex to the invader. The CRISPR-Cas types I and III use the Cas6 protein to generate mature crRNAs. Here, we show that the Cas6 protein is necessary for crRNA production but that additional Cas proteins that form a CRISPR-associated complex for antiviral defense (Cascade)-like complex are needed for crRNA stability in the CRISPR-Cas type I-B system in Haloferax volcanii in vivo. Deletion of the cas6 gene results in the loss of mature crRNAs and interference. However, cells that have the complete cas gene cluster (cas1-8b) removed and are transformed with the cas6 gene are not able to produce and stably maintain mature crRNAs. crRNA production and stability is rescued only if cas5, -6, and -7 are present. Mutational analysis of the cas6 gene reveals three amino acids (His-41, Gly-256, and Gly-258) that are essential for pre-crRNA cleavage, whereas the mutation of two amino acids (Ser-115 and Ser-224) leads to an increase of crRNA amounts. This is the first systematic in vivo analysis of Cas6 protein variants. In addition, we show that the H. volcanii I-B system contains a Cascade-like complex with a Cas7, Cas5, and Cas6 core that protects the crRNA.
Keywords: Archaea; CRISPR/Cas; Cas6; Haloferax volcanii; Microbiology; Molecular Biology; Molecular Genetics; Protein Complexes; Type I-B; crRNA.
Figures



Similar articles
-
An active immune defense with a minimal CRISPR (clustered regularly interspaced short palindromic repeats) RNA and without the Cas6 protein.J Biol Chem. 2015 Feb 13;290(7):4192-201. doi: 10.1074/jbc.M114.617506. Epub 2014 Dec 15. J Biol Chem. 2015. PMID: 25512373 Free PMC article.
-
Essential requirements for the detection and degradation of invaders by the Haloferax volcanii CRISPR/Cas system I-B.RNA Biol. 2013 May;10(5):865-74. doi: 10.4161/rna.24282. Epub 2013 Apr 17. RNA Biol. 2013. PMID: 23594992 Free PMC article.
-
Characterization of CRISPR RNA biogenesis and Cas6 cleavage-mediated inhibition of a provirus in the haloarchaeon Haloferax mediterranei.J Bacteriol. 2013 Feb;195(4):867-75. doi: 10.1128/JB.01688-12. Epub 2012 Dec 14. J Bacteriol. 2013. PMID: 23243301 Free PMC article.
-
Approaches to study CRISPR RNA biogenesis and the key players involved.Methods. 2020 Feb 1;172:12-26. doi: 10.1016/j.ymeth.2019.07.015. Epub 2019 Jul 17. Methods. 2020. PMID: 31325492 Review.
-
Biogenesis pathways of RNA guides in archaeal and bacterial CRISPR-Cas adaptive immunity.FEMS Microbiol Rev. 2015 May;39(3):428-41. doi: 10.1093/femsre/fuv023. Epub 2015 May 19. FEMS Microbiol Rev. 2015. PMID: 25994611 Free PMC article. Review.
Cited by
-
An archaeal CRISPR type III-B system exhibiting distinctive RNA targeting features and mediating dual RNA and DNA interference.Nucleic Acids Res. 2015 Jan;43(1):406-17. doi: 10.1093/nar/gku1302. Epub 2014 Dec 10. Nucleic Acids Res. 2015. PMID: 25505143 Free PMC article.
-
An archaeal Cas3 protein facilitates rapid recovery from DNA damage.Microlife. 2023 Feb 9;4:uqad007. doi: 10.1093/femsml/uqad007. eCollection 2023. Microlife. 2023. PMID: 37223740 Free PMC article.
-
Detection and characterization of clustered regularly interspaced short palindromic repeat-associated endoribonuclease gene variants in Vibrio parahaemolyticus isolated from seafoods and environment.Vet World. 2019 May;12(5):689-695. doi: 10.14202/vetworld.2019.689-695. Epub 2019 May 21. Vet World. 2019. PMID: 31327905 Free PMC article.
-
The switch complex ArlCDE connects the chemotaxis system and the archaellum.Mol Microbiol. 2020 Sep;114(3):468-479. doi: 10.1111/mmi.14527. Epub 2020 Jun 8. Mol Microbiol. 2020. PMID: 32416640 Free PMC article.
-
Expansion of the CRISPR/Cas Genome-Sculpting Toolbox: Innovations, Applications and Challenges.Mol Diagn Ther. 2021 Jan;25(1):41-57. doi: 10.1007/s40291-020-00500-8. Epub 2020 Nov 13. Mol Diagn Ther. 2021. PMID: 33185860 Review.
References
-
- Al-Attar S., Westra E. R., van der Oost J., Brouns S. J. (2011) Clustered regularly interspaced short palindromic repeats (CRISPRs). The hallmark of an ingenious antiviral defense mechanism in prokaryotes. Biol. Chem. 392, 277–289 - PubMed
-
- Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D. A., Horvath P. (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 - PubMed
-
- Bhaya D., Davison M., Barrangou R. (2011) CRISPR-Cas systems in bacteria and archaea. Versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. 45, 273–297 - PubMed
-
- Garneau J. E., Dupuis M. È., Villion M., Romero D. A., Barrangou R., Boyaval P., Fremaux C., Horvath P., Magadán A. H., Moineau S. (2010) The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67–71 - PubMed
-
- Garrett R. A., Vestergaard G., Shah S. A. (2011) Archaeal CRISPR-based immune systems. Exchangeable functional modules. Trends Microbiol. 19, 549–556 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

