BuT2 is a member of the third major group of hAT transposons and is involved in horizontal transfer events in the genus Drosophila
- PMID: 24459285
- PMCID: PMC3942097
- DOI: 10.1093/gbe/evu017
BuT2 is a member of the third major group of hAT transposons and is involved in horizontal transfer events in the genus Drosophila
Abstract
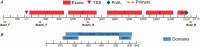
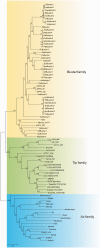
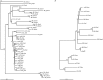
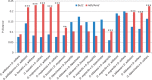
The hAT superfamily comprises a large and diverse array of DNA transposons found in all supergroups of eukaryotes. Here we characterized the Drosophila buzzatii BuT2 element and found that it harbors a five-exon gene encoding a 643-aa putatively functional transposase. A phylogeny built with 85 hAT transposases yielded, in addition to the two major groups already described, Ac and Buster, a third one comprising 20 sequences that includes BuT2, Tip100, hAT-4_BM, and RP-hAT1. This third group is here named Tip. In addition, we studied the phylogenetic distribution and evolution of BuT2 by in silico searches and molecular approaches. Our data revealed BuT2 was, most often, vertically transmitted during the evolution of genus Drosophila being lost independently in several species. Nevertheless, we propose the occurrence of three horizontal transfer events to explain its distribution and conservation among species. Another aspect of BuT2 evolution and life cycle is the presence of short related sequences, which contain similar 5' and 3' regions, including the terminal inverted repeats. These sequences that can be considered as miniature inverted repeat transposable elements probably originated by internal deletion of complete copies and show evidences of recent mobilization.
Keywords: Drosophila; MITE; hAT; horizontal transfer; transposase.
Figures

Similar articles
-
Characterization of Herves-like transposable elements (hATs) in Drosophila species and their evolutionary scenario.Insect Mol Biol. 2019 Oct;28(5):616-627. doi: 10.1111/imb.12577. Epub 2019 Mar 12. Insect Mol Biol. 2019. PMID: 30793407
-
Characterization of new hAT transposable elements in 12 Drosophila genomes.Genetica. 2009 Jan;135(1):67-75. doi: 10.1007/s10709-008-9259-5. Epub 2008 Mar 14. Genetica. 2009. PMID: 18340538
-
Structure and evolution of the hAT transposon superfamily.Genetics. 2001 Jul;158(3):949-57. doi: 10.1093/genetics/158.3.949. Genetics. 2001. PMID: 11454746 Free PMC article.
-
Horizontal transfer of P elements and other short inverted repeat transposons.Genetica. 1992;86(1-3):275-86. doi: 10.1007/BF00133726. Genetica. 1992. PMID: 1334912 Review.
-
The hAT family: a versatile transposon group common to plants, fungi, animals, and man.Chromosoma. 2001 Apr;110(1):1-9. doi: 10.1007/s004120000118. Chromosoma. 2001. PMID: 11398971 Review.
Cited by
-
Unexpected invasion of miniature inverted-repeat transposable elements in viral genomes.Mob DNA. 2018 Jun 18;9:19. doi: 10.1186/s13100-018-0125-4. eCollection 2018. Mob DNA. 2018. PMID: 29946369 Free PMC article.
-
Interpopulation variation of transposable elements of the hAT superfamily in Drosophila willistoni (Diptera: Drosophilidae): in-situ approach.Genet Mol Biol. 2022 Mar 16;45(2):e20210287. doi: 10.1590/1678-4685-GMB-2021-0287. eCollection 2022. Genet Mol Biol. 2022. PMID: 35297941 Free PMC article.
-
Characterization of a novel Helitron family in insect genomes: insights into classification, evolution and horizontal transfer.Mob DNA. 2019 May 31;10:25. doi: 10.1186/s13100-019-0165-4. eCollection 2019. Mob DNA. 2019. PMID: 31164927 Free PMC article.
-
Horizontal Gene Transfer: From Evolutionary Flexibility to Disease Progression.Front Cell Dev Biol. 2020 May 19;8:229. doi: 10.3389/fcell.2020.00229. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 32509768 Free PMC article. Review.
-
Two hAT transposon genes were transferred from Brassicaceae to broomrapes and are actively expressed in some recipients.Sci Rep. 2016 Jul 25;6:30192. doi: 10.1038/srep30192. Sci Rep. 2016. PMID: 27452947 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

