JAK/STAT Signalling in Huntington's Disease Immune Cells
- PMID: 24459609
- PMCID: PMC3871417
- DOI: 10.1371/currents.hd.5791c897b5c3bebeed93b1d1da0c0648
JAK/STAT Signalling in Huntington's Disease Immune Cells
Abstract
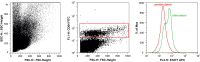
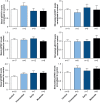
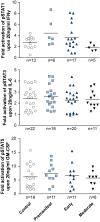
Huntington's disease (HD) is an inherited neurodegenerative disorder caused by a CAG repeat expansion in the huntingtin (HTT) gene. Both central and peripheral innate immune activation have been described as features of the disease. Isolated human HD monocytes have been shown to produce more cytokines upon LPS stimulation compared to control monocytes. Understanding alterations in the signalling cascades responsible and activated by this increase in pro-inflammatory cytokine production is crucial in understanding the molecular basis of this phenomenon. Here we investigated the signalling cascade most commonly activated by pro-inflammatory cytokines such as IL-6 - the JAK/STAT signalling cascade. Using flow cytometry, we show that one out of three key transcription factors activated by JAK/STAT signalling is altered in primary human HD innate immune cells, suggesting that this pathway may only play a minor, additive role in the immune cell dysfunction in HD.
Figures

References
-
- Bates G, Harper PS, Jones L, editors (2002) Huntington's Disease. Oxford: Oxford University Press.
-
- Li SH, Schilling G, Young WS, Li XJ, Margolis RL, et al. (1993) Huntington's disease gene (IT15) is widely expressed in human and rat tissues. Neuron 11: 985-993. - PubMed
-
- van der Burg JM, Björkqvist M, Brundin P (2009) Beyond the brain: widespread pathology in Huntington's disease. Lancet Neurol 8: 765-774. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

