Lung microbiome for clinicians. New discoveries about bugs in healthy and diseased lungs
- PMID: 24460444
- PMCID: PMC3972985
- DOI: 10.1513/AnnalsATS.201310-339FR
Lung microbiome for clinicians. New discoveries about bugs in healthy and diseased lungs
Abstract
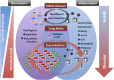
Microbes are readily cultured from epithelial surfaces of the skin, mouth, and colon. In the last 10 years, culture-independent DNA-based techniques demonstrated that much more complex microbial communities reside on most epithelial surfaces; this includes the lower airways, where bacterial culture had failed to reliably demonstrate resident bacteria. Exposure to a diverse bacterial environment is important for adequate immunological development. The most common microbes found in the lower airways are also found in the upper airways. Increasing abundance of oral characteristic taxa is associated with increased inflammatory cells and exhaled nitric oxide, suggesting that the airway microbiome induces an immunological response in the lung. Furthermore, rhinovirus infection leads to outgrowth of Haemophilus in patients with chronic obstructive pulmonary disease, and human immunodeficiency virus-infected subjects have more Tropheryma whipplei in the lower airway, suggesting a bidirectional interaction in which the host immune defenses also influence the microbial niche. Quantitative and/or qualitative changes in the lung microbiome may be relevant for disease progression and exacerbations in a number of pulmonary diseases. Future investigations with longitudinal follow-up to understand the dynamics of the lung microbiome may lead to the development of new therapeutic targets.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical