A founder mutation in PET100 causes isolated complex IV deficiency in Lebanese individuals with Leigh syndrome
- PMID: 24462369
- PMCID: PMC3928654
- DOI: 10.1016/j.ajhg.2013.12.015
A founder mutation in PET100 causes isolated complex IV deficiency in Lebanese individuals with Leigh syndrome
Abstract
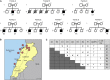
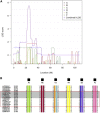
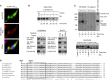
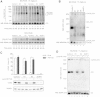
Leigh syndrome (LS) is a severe neurodegenerative disorder with characteristic bilateral lesions, typically in the brainstem and basal ganglia. It usually presents in infancy and is genetically heterogeneous, but most individuals with mitochondrial complex IV (or cytochrome c oxidase) deficiency have mutations in the biogenesis factor SURF1. We studied eight complex IV-deficient LS individuals from six families of Lebanese origin. They differed from individuals with SURF1 mutations in having seizures as a prominent feature. Complementation analysis suggested they had mutation(s) in the same gene but targeted massively parallel sequencing (MPS) of 1,034 genes encoding known mitochondrial proteins failed to identify a likely candidate. Linkage and haplotype analyses mapped the location of the gene to chromosome 19 and targeted MPS of the linkage region identified a homozygous c.3G>C (p.Met1?) mutation in C19orf79. Abolishing the initiation codon could potentially still allow initiation at a downstream methionine residue but we showed that this would not result in a functional protein. We confirmed that mutation of this gene was causative by lentiviral-mediated phenotypic correction. C19orf79 was recently renamed PET100 and predicted to encode a complex IV biogenesis factor. We showed that it is located in the mitochondrial inner membrane and forms a ∼300 kDa subcomplex with complex IV subunits. Previous proteomic analyses of mitochondria had overlooked PET100 because its small size was below the cutoff for annotating bona fide proteins. The mutation was estimated to have arisen at least 520 years ago, explaining how the families could have different religions and different geographic origins within Lebanon.
Copyright © 2014 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Zhu Z., Yao J., Johns T., Fu K., De Bie I., Macmillan C., Cuthbert A.P., Newbold R.F., Wang J., Chevrette M. SURF1, encoding a factor involved in the biogenesis of cytochrome c oxidase, is mutated in Leigh syndrome. Nat. Genet. 1998;20:337–343. - PubMed
-
- Kirby D.M., Thorburn D.R. Approaches to finding the molecular basis of mitochondrial oxidative phosphorylation disorders. Twin Res. Hum. Genet. 2008;11:395–411. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- RC2 HL102926/HL/NHLBI NIH HHS/United States
- HL-102924/HL/NHLBI NIH HHS/United States
- RC2 HL102924/HL/NHLBI NIH HHS/United States
- HL-102926/HL/NHLBI NIH HHS/United States
- HL-102925/HL/NHLBI NIH HHS/United States
- RC2 HL103010/HL/NHLBI NIH HHS/United States
- HL-102923/HL/NHLBI NIH HHS/United States
- RC2 HL102923/HL/NHLBI NIH HHS/United States
- UC2 HL102926/HL/NHLBI NIH HHS/United States
- UC2 HL103010/HL/NHLBI NIH HHS/United States
- HL-103010/HL/NHLBI NIH HHS/United States
- UC2 HL102923/HL/NHLBI NIH HHS/United States
- UC2 HL102924/HL/NHLBI NIH HHS/United States
- RC2 HL102925/HL/NHLBI NIH HHS/United States
- UC2 HL102925/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

