Aberrant expression of SALL4 in acute B cell lymphoblastic leukemia: mechanism, function, and implication for a potential novel therapeutic target
- PMID: 24463278
- PMCID: PMC4135469
- DOI: 10.1016/j.exphem.2014.01.005
Aberrant expression of SALL4 in acute B cell lymphoblastic leukemia: mechanism, function, and implication for a potential novel therapeutic target
Abstract
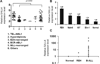
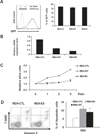
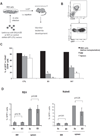
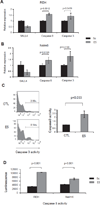
Treatment for high-risk pediatric and adult acute B cell lymphoblastic leukemia (B-ALL) remains challenging. Exploring novel pathways in B-ALL could lead to new therapy. Our previous study has shown that stem cell factor SALL4 is aberrantly expressed in B-ALL, but its functional roles and the mechanism that accounts for its upregulation in B-ALL remain unexplored. To address this question, we first surveyed the existing B-ALL cell lines and primary patient samples for SALL4 expression. We then selected the B-ALL cell lines with the highest SALL4 expression for functional studies. RNA interference was used to downregulate SALL4 expression in these cell lines. When compared with control cells, SALL4 knockdown cells exhibited decreased cell proliferation, increased apoptosis in vitro, and decreased engraftment in a xenotransplant model in vivo. Gene expression analysis showed that in SALL4 knockdown B-ALL cells, multiple caspase members involved in cell apoptosis pathway were upregulated. Next, we explored the mechanisms of aberrant SALL4 expression in B-ALL. We found that hypomethylation of the SALL4 CpG islands was correlated with its high expression. Furthermore, treatment of low SALL4-expressing B-ALL cell lines with DNA methylation inhibitor led to demethylation of the SALL4 CpG and increased SALL4 expression. In summary, to our knowledge, we are the first to show that the aberrant expression of SALL4 in B-ALL is associated with hypomethylation, and that SALL4 plays a key role in B-ALL cell survival and could be a potential novel target in B-ALL treatment.
Copyright © 2014 ISEH - Society for Hematology and Stem Cells. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
No financial interest/relationships with financial interest relating to the topic of this article have been declared.
Figures



References
-
- Pui CH, Relling MV, Downing JR. Acute lymphoblastic leukemia. New Engl J Med. 2004;350:1535–1548. - PubMed
-
- Medvedovic J, Ebert A, Tagoh H, Busslinger M. Pax5: a master regulator of B cell development and leukemogenesis. Adv Immunol. 2011;111:179–206. - PubMed
-
- Kohlhase J, Heinrich M, Schubert L, et al. Okihiro syndrome is caused by SALL4 mutations. Hum Mol Genet. 2002;11:2979–2987. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

