Crystal structure of a Schistosoma mansoni septin reveals the phenomenon of strand slippage in septins dependent on the nature of the bound nucleotide
- PMID: 24464615
- PMCID: PMC3953292
- DOI: 10.1074/jbc.M113.525352
Crystal structure of a Schistosoma mansoni septin reveals the phenomenon of strand slippage in septins dependent on the nature of the bound nucleotide
Abstract
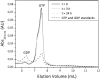
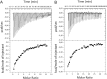
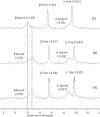
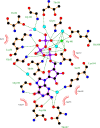
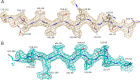
Septins are filament-forming GTP-binding proteins involved in important cellular events, such as cytokinesis, barrier formation, and membrane remodeling. Here, we present two crystal structures of the GTPase domain of a Schistosoma mansoni septin (SmSEPT10), one bound to GDP and the other to GTP. The structures have been solved at an unprecedented resolution for septins (1.93 and 2.1 Å, respectively), which has allowed for unambiguous structural assignment of regions previously poorly defined. Consequently, we provide a reliable model for functional interpretation and a solid foundation for future structural studies. Upon comparing the two complexes, we observe for the first time the phenomenon of a strand slippage in septins. Such slippage generates a front-back communication mechanism between the G and NC interfaces. These data provide a novel mechanistic framework for the influence of nucleotide binding to the GTPase domain, opening new possibilities for the study of the dynamics of septin filaments.
Keywords: Conformational Change; Crystal Structure; Filament; GTPase; GTPase Domain; Protein Conformation; Protein Structure; Schistosoma; Septin; X-ray Crystallography.
Figures






Similar articles
-
Revisiting SEPT7 and the slippage of β-strands in the septin family.J Struct Biol. 2019 Jul 1;207(1):67-73. doi: 10.1016/j.jsb.2019.04.015. Epub 2019 Apr 19. J Struct Biol. 2019. PMID: 31009756
-
In silico docking of forchlorfenuron (FCF) to septins suggests that FCF interferes with GTP binding.PLoS One. 2014 May 2;9(5):e96390. doi: 10.1371/journal.pone.0096390. eCollection 2014. PLoS One. 2014. PMID: 24787956 Free PMC article.
-
Biophysical dissection of schistosome septins: Insights into oligomerization and membrane binding.Biochimie. 2016 Dec;131:96-105. doi: 10.1016/j.biochi.2016.09.014. Epub 2016 Sep 26. Biochimie. 2016. PMID: 27687162
-
A biochemical view on the septins, a less known component of the cytoskeleton.Biol Chem. 2022 Nov 25;404(1):1-13. doi: 10.1515/hsz-2022-0263. Print 2023 Jan 27. Biol Chem. 2022. PMID: 36423333 Review.
-
The evolution, complex structures and function of septin proteins.Cell Mol Life Sci. 2009 Oct;66(20):3309-23. doi: 10.1007/s00018-009-0087-2. Epub 2009 Jul 14. Cell Mol Life Sci. 2009. PMID: 19597764 Free PMC article. Review.
Cited by
-
Septin Mutations in Human Cancers.Front Cell Dev Biol. 2016 Nov 9;4:122. doi: 10.3389/fcell.2016.00122. eCollection 2016. Front Cell Dev Biol. 2016. PMID: 27882315 Free PMC article. Review.
-
High pressure 31P NMR spectroscopy on guanine nucleotides.J Biomol NMR. 2017 Jan;67(1):1-13. doi: 10.1007/s10858-016-0079-0. Epub 2016 Dec 23. J Biomol NMR. 2017. PMID: 28012125
-
Effects of Bni5 Binding on Septin Filament Organization.J Mol Biol. 2016 Dec 4;428(24 Pt B):4962-4980. doi: 10.1016/j.jmb.2016.10.024. Epub 2016 Oct 30. J Mol Biol. 2016. PMID: 27806918 Free PMC article.
-
A FRET-based method for monitoring septin polymerization and binding of septin-associated proteins.Methods Cell Biol. 2016;136:35-56. doi: 10.1016/bs.mcb.2016.03.024. Epub 2016 Jun 14. Methods Cell Biol. 2016. PMID: 27473902 Free PMC article.
-
The Mammalian Septin Interactome.Front Cell Dev Biol. 2017 Feb 7;5:3. doi: 10.3389/fcell.2017.00003. eCollection 2017. Front Cell Dev Biol. 2017. PMID: 28224124 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

