The crystal structure of TDP-43 RRM1-DNA complex reveals the specific recognition for UG- and TG-rich nucleic acids
- PMID: 24464995
- PMCID: PMC3985631
- DOI: 10.1093/nar/gkt1407
The crystal structure of TDP-43 RRM1-DNA complex reveals the specific recognition for UG- and TG-rich nucleic acids
Abstract
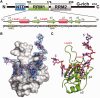
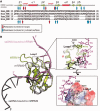
TDP-43 is an important pathological protein that aggregates in the diseased neuronal cells and is linked to various neurodegenerative disorders. In normal cells, TDP-43 is primarily an RNA-binding protein; however, how the dimeric TDP-43 binds RNA via its two RNA recognition motifs, RRM1 and RRM2, is not clear. Here we report the crystal structure of human TDP-43 RRM1 in complex with a single-stranded DNA showing that RRM1 binds the nucleic acid extensively not only by the conserved β-sheet residues but also by the loop residues. Mutational and biochemical assays further reveal that both RRMs in TDP-43 dimers participate in binding of UG-rich RNA or TG-rich DNA with RRM1 playing a dominant role and RRM2 playing a supporting role. Moreover, RRM1 of the amyotrophic lateral sclerosis-linked mutant D169G binds DNA as efficiently as the wild type; nevertheless, it is more resistant to thermal denaturation, suggesting that the resistance to degradation is likely linked to TDP-43 proteinopathies. Taken together all the data, we suggest a model showing that the two RRMs in each protomer of TDP-43 homodimer work together in RNA binding and thus the dimeric TDP-43 recognizes long clusters of UG-rich RNA to achieve high affinity and specificity.
Figures




References
-
- Wang I-F, Wu L-S, Shen C-KJ. TDP-43: an emerging new player in neurodegenerative diseases. Trends Mol. Med. 2008;14:479–485. - PubMed
-
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. - PubMed
-
- Arai T, Hasegawa M, Akiyama H, Ikeda K, Nonaka T, Mori H, Mann D, Tsuchiya K, Yoshida M, Hashizume Y, et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Comm. 2006;351:602–611. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

