Cytokinins
- PMID: 24465173
- PMCID: PMC3894907
- DOI: 10.1199/tab.0168
Cytokinins
Abstract
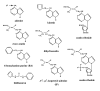
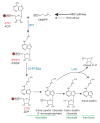
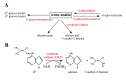
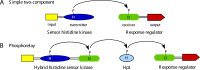
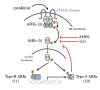
Cytokinins are N (6) substituted adenine derivatives that affect many aspects of plant growth and development, including cell division, shoot initiation and growth, leaf senescence, apical dominance, sink/source relationships, nutrient uptake, phyllotaxis, and vascular, gametophyte, and embryonic development, as well as the response to biotic and abiotic factors. Molecular genetic studies in Arabidopsis have helped elucidate the mechanisms underlying the function of this phytohormone in plants. Here, we review our current understanding of cytokinin biosynthesis and signaling in Arabidopsis, the latter of which is similar to bacterial two-component phosphorelays. We discuss the perception of cytokinin by the ER-localized histidine kinase receptors, the role of the AHPs in mediating the transfer of the phosphoryl group from the receptors to the response regulators (ARRs), and finally the role of the large ARR family in cytokinin function. The identification and genetic manipulation of the genes involved in cytokinin metabolism and signaling have helped illuminate the roles of cytokinins in Arabidopsis. We discuss these diverse roles, and how other signaling pathways influence cytokinin levels and sensitivity though modulation of the expression of cytokinin signaling and metabolic genes.
Figures
References
-
- Alvarez S., Marsh E.L., Schroeder S.G., Schachtman D.P. Metabolomic and proteomic changes in the xylem sap of maize under drought. Plant Cell Environ. 2008;31:325–340. - PubMed
-
- Anantharaman V., Aravind L. The CHASE domain: a predicted ligand-binding module in plant cytokinin receptors and other eukaryotic and bacterial receptors. Trends Biochem. Sci. 2001;26:579–582. - PubMed
-
- Appleby J.L., Parkinson J.S., Bourret R.B. Signal transduction via the multi-step phosphorelay: not necessarily a road less traveled. Cell. 1996;86:845–848. - PubMed
-
- Argueso C.T., Ferreira F.J., Kieber J.J. Environmental perception avenues: the interaction of cytokinin and environmental response pathways. Plant Cell Environ. 2009;32:1147–1160. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
