Detection of host-derived sphingosine by Pseudomonas aeruginosa is important for survival in the murine lung
- PMID: 24465209
- PMCID: PMC3900636
- DOI: 10.1371/journal.ppat.1003889
Detection of host-derived sphingosine by Pseudomonas aeruginosa is important for survival in the murine lung
Abstract
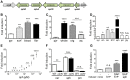
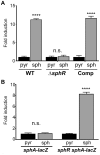
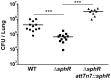
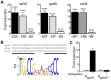
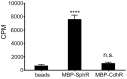
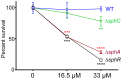
Pseudomonas aeruginosa is a common environmental bacterium that is also a significant opportunistic pathogen, particularly of the human lung. We must understand how P. aeruginosa responds to the lung environment in order to identify the regulatory changes that bacteria use to establish and maintain infections. The P. aeruginosa response to pulmonary surfactant was used as a model to identify transcripts likely induced during lung infection. The most highly induced transcript in pulmonary surfactant, PA5325 (sphA), is regulated by an AraC-family transcription factor, PA5324 (SphR). We found that sphA was specifically induced by sphingosine in an SphR-dependent manner, and also via metabolism of sphingomyelin, ceramide, or sphingoshine-1-phosphate to sphingosine. These sphingolipids not only play a structural role in lipid membranes, but some are also intracellular and intercellular signaling molecules important in normal eukaryotic cell functions as well as orchestrating immune responses. The members of the SphR transcriptome were identified by microarray analyses, and DNA binding assays showed specific interaction of these promoters with SphR, which enabled us to determine the consensus SphR binding site. SphR binding to DNA was modified by sphingosine and we used labeled sphingosine to demonstrate direct binding of sphingosine by SphR. Deletion of sphR resulted in reduced bacterial survival during mouse lung infection. In vitro experiments show that deletion of sphR increases sensitivity to the antimicrobial effects of sphingosine which could, in part, explain the in vivo phenotype. This is the first identification of a sphingosine-responsive transcription factor in bacteria. We predict that SphR transcriptional regulation may be important in response to many sites of infection in eukaryotes and the presence of homologous transcription factors in other pathogens suggests that sphingosine detection is not limited to P. aeruginosa.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
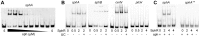

Similar articles
-
Molecular mechanism for sphingosine-induced Pseudomonas ceramidase expression through the transcriptional regulator SphR.Sci Rep. 2016 Dec 12;6:38797. doi: 10.1038/srep38797. Sci Rep. 2016. PMID: 27941831 Free PMC article.
-
Genetic deletion of Sphk2 confers protection against Pseudomonas aeruginosa mediated differential expression of genes related to virulent infection and inflammation in mouse lung.BMC Genomics. 2019 Dec 16;20(1):984. doi: 10.1186/s12864-019-6367-9. BMC Genomics. 2019. PMID: 31842752 Free PMC article.
-
Requirement of the Pseudomonas aeruginosa CbrA sensor kinase for full virulence in a murine acute lung infection model.Infect Immun. 2014 Mar;82(3):1256-67. doi: 10.1128/IAI.01527-13. Epub 2013 Dec 30. Infect Immun. 2014. PMID: 24379284 Free PMC article.
-
Pseudomonas aeruginosa: breaking down barriers.Curr Genet. 2016 Feb;62(1):109-13. doi: 10.1007/s00294-015-0522-x. Epub 2015 Sep 25. Curr Genet. 2016. PMID: 26407972 Free PMC article. Review.
-
Airway immunometabolites fuel Pseudomonas aeruginosa infection.Respir Res. 2020 Dec 10;21(1):326. doi: 10.1186/s12931-020-01591-x. Respir Res. 2020. PMID: 33302964 Free PMC article. Review.
Cited by
-
Evidence and Role for Bacterial Mucin Degradation in Cystic Fibrosis Airway Disease.PLoS Pathog. 2016 Aug 22;12(8):e1005846. doi: 10.1371/journal.ppat.1005846. eCollection 2016 Aug. PLoS Pathog. 2016. PMID: 27548479 Free PMC article.
-
The AraC-Type Transcriptional Regulator GliR (PA3027) Activates Genes of Glycerolipid Metabolism in Pseudomonas aeruginosa.Int J Mol Sci. 2021 May 11;22(10):5066. doi: 10.3390/ijms22105066. Int J Mol Sci. 2021. PMID: 34064685 Free PMC article.
-
Sphingosine kills bacteria by binding to cardiolipin.J Biol Chem. 2020 May 29;295(22):7686-7696. doi: 10.1074/jbc.RA119.012325. Epub 2020 Apr 23. J Biol Chem. 2020. PMID: 32327486 Free PMC article.
-
Intestinal Long-Chain Fatty Acids Act as a Direct Signal To Modulate Expression of the Salmonella Pathogenicity Island 1 Type III Secretion System.mBio. 2016 Feb 16;7(1):e02170-15. doi: 10.1128/mBio.02170-15. mBio. 2016. PMID: 26884427 Free PMC article.
-
The Pseudomonas aeruginosa sphBC genes are important for growth in the presence of sphingosine by promoting sphingosine metabolism.bioRxiv [Preprint]. 2024 Sep 3:2024.09.03.611043. doi: 10.1101/2024.09.03.611043. bioRxiv. 2024. Update in: Microbiology (Reading). 2025 Jan;171(1). doi: 10.1099/mic.0.001520. PMID: 39282278 Free PMC article. Updated. Preprint.
References
-
- Lieberman D, Lieberman D (2003) Pseudomonal infections in patients with COPD: epidemiology and management. Am J Respir Med 2: 459–468. - PubMed
-
- Chastre J, Fagon JY (2002) Ventilator-associated pneumonia. Am J Respir Crit Care Med 165: 867–903. - PubMed
-
- Crouch Brewer S, Wunderink RG, Jones CB, Leeper KV Jr (1996) Ventilator-associated pneumonia due to Pseudomonas aeruginosa . Chest 109: 1019–1029. - PubMed
-
- Burns JL, Emerson J, Stapp JR, Yim DL, Krzewinski J, et al. (1998) Microbiology of sputum from patients at cystic fibrosis centers in the United States. Clin Infect Dis 27: 158–163. - PubMed
-
- Briesacher BA, Quittner AL, Fouayzi H, Zhang J, Swensen A (2011) Nationwide trends in the medical care costs of privately insured patients with cystic fibrosis (CF), 2001–2007. Pediatr Pulmonol 46: 770–776. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

