The SET domain proteins SUVH2 and SUVH9 are required for Pol V occupancy at RNA-directed DNA methylation loci
- PMID: 24465213
- PMCID: PMC3898904
- DOI: 10.1371/journal.pgen.1003948
The SET domain proteins SUVH2 and SUVH9 are required for Pol V occupancy at RNA-directed DNA methylation loci
Abstract
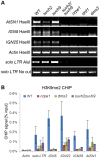
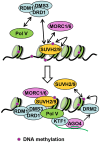
RNA-directed DNA methylation (RdDM) is required for transcriptional silencing of transposons and other DNA repeats in Arabidopsis thaliana. Although previous research has demonstrated that the SET domain-containing SU(VAR)3-9 homologs SUVH2 and SUVH9 are involved in the RdDM pathway, the underlying mechanism remains unknown. Our results indicated that SUVH2 and/or SUVH9 not only interact with the chromatin-remodeling complex termed DDR (DMS3, DRD1, and RDM1) but also with the newly characterized complex composed of two conserved Microrchidia (MORC) family proteins, MORC1 and MORC6. The effect of suvh2suvh9 on Pol IV-dependent siRNA accumulation and DNA methylation is comparable to that of the Pol V mutant nrpe1 and the DDR complex mutant dms3, suggesting that SUVH2 and SUVH9 are functionally associated with RdDM. Our CHIP assay demonstrated that SUVH2 and SUVH9 are required for the occupancy of Pol V at RdDM loci and facilitate the production of Pol V-dependent noncoding RNAs. Moreover, SUVH2 and SUVH9 are also involved in the occupancy of DMS3 at RdDM loci. The putative catalytic active site in the SET domain of SUVH2 is dispensable for the function of SUVH2 in RdDM and H3K9 dimethylation. We propose that SUVH2 and SUVH9 bind to methylated DNA and facilitate the recruitment of Pol V to RdDM loci by associating with the DDR complex and the MORC complex.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
SUVH2 and SUVH9 Couple Two Essential Steps for Transcriptional Gene Silencing in Arabidopsis.Mol Plant. 2016 Aug 1;9(8):1156-1167. doi: 10.1016/j.molp.2016.05.006. Epub 2016 May 20. Mol Plant. 2016. PMID: 27216319
-
SRA- and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation.Nature. 2014 Mar 6;507(7490):124-128. doi: 10.1038/nature12931. Epub 2014 Jan 22. Nature. 2014. PMID: 24463519 Free PMC article.
-
Two Components of the RNA-Directed DNA Methylation Pathway Associate with MORC6 and Silence Loci Targeted by MORC6 in Arabidopsis.PLoS Genet. 2016 May 12;12(5):e1006026. doi: 10.1371/journal.pgen.1006026. eCollection 2016 May. PLoS Genet. 2016. PMID: 27171427 Free PMC article.
-
The RNAs of RNA-directed DNA methylation.Biochim Biophys Acta Gene Regul Mech. 2017 Jan;1860(1):140-148. doi: 10.1016/j.bbagrm.2016.08.004. Epub 2016 Aug 10. Biochim Biophys Acta Gene Regul Mech. 2017. PMID: 27521981 Free PMC article. Review.
-
Non-coding RNA transcription and RNA-directed DNA methylation in Arabidopsis.Mol Plant. 2014 Sep;7(9):1406-1414. doi: 10.1093/mp/ssu075. Epub 2014 Jun 25. Mol Plant. 2014. PMID: 24966349 Review.
Cited by
-
Arabidopsis RNA Polymerases IV and V Are Required To Establish H3K9 Methylation, but Not Cytosine Methylation, on Geminivirus Chromatin.J Virol. 2016 Jul 27;90(16):7529-7540. doi: 10.1128/JVI.00656-16. Print 2016 Aug 15. J Virol. 2016. PMID: 27279611 Free PMC article.
-
More than meets the eye? Factors that affect target selection by plant miRNAs and heterochromatic siRNAs.Curr Opin Plant Biol. 2015 Oct;27:118-24. doi: 10.1016/j.pbi.2015.06.012. Epub 2015 Jul 31. Curr Opin Plant Biol. 2015. PMID: 26246393 Free PMC article. Review.
-
Transcriptional gene silencing by Arabidopsis microrchidia homologues involves the formation of heteromers.Proc Natl Acad Sci U S A. 2014 May 20;111(20):7474-9. doi: 10.1073/pnas.1406611111. Epub 2014 May 5. Proc Natl Acad Sci U S A. 2014. PMID: 24799676 Free PMC article.
-
Epigenetic regulation of plant immunity: from chromatin codes to plant disease resistance.aBIOTECH. 2023 Mar 17;4(2):124-139. doi: 10.1007/s42994-023-00101-z. eCollection 2023 Jun. aBIOTECH. 2023. PMID: 37581024 Free PMC article. Review.
-
Genome-wide identification and expression profiling of SET DOMAIN GROUP family in Dendrobium catenatum.BMC Plant Biol. 2020 Jan 28;20(1):40. doi: 10.1186/s12870-020-2244-6. BMC Plant Biol. 2020. PMID: 31992218 Free PMC article.
References
-
- Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJ (2009) RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol 21: 367–376. - PubMed
-
- Mirouze M, Reinders J, Bucher E, Nishimura T, Schneeberger K, et al. (2009) Selective epigenetic control of retrotransposition in Arabidopsis. Nature 461: 427–430. - PubMed
-
- Tsukahara S, Kobayashi A, Kawabe A, Mathieu O, Miura A, et al. (2009) Bursts of retrotransposition reproduced in Arabidopsis. Nature 461: 423–426. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

