Arabidopsis AL PHD-PRC1 complexes promote seed germination through H3K4me3-to-H3K27me3 chromatin state switch in repression of seed developmental genes
- PMID: 24465219
- PMCID: PMC3900384
- DOI: 10.1371/journal.pgen.1004091
Arabidopsis AL PHD-PRC1 complexes promote seed germination through H3K4me3-to-H3K27me3 chromatin state switch in repression of seed developmental genes
Abstract
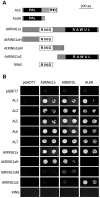
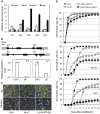
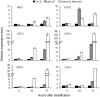
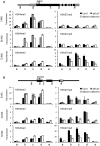
Seed germination and subsequent seedling growth define crucial steps for entry into the plant life cycle. For those events to take place properly, seed developmental genes need to be silenced whereas vegetative growth genes are activated. Chromatin structure is generally known to play crucial roles in gene transcription control. However, the transition between active and repressive chromatin states during seed germination is still poorly characterized and the underlying molecular mechanisms remain largely unknown. Here we identified the Arabidopsis PHD-domain H3K4me3-binding ALFIN1-like proteins (ALs) as novel interactors of the Polycomb Repressive Complex 1 (PRC1) core components AtBMI1b and AtRING1a. The interactions were confirmed by diverse in vitro and in vivo assays and were shown to require the AL6 N-terminus containing PAL domain conserved in the AL family proteins and the AtRING1a C-terminus containing RAWUL domain conserved in animal and plant PRC1 ring-finger proteins (including AtRNIG1a/b and AtBMI1a/b). By T-DNA insertion mutant analysis, we found that simultaneous loss of AL6 and AL7 as well as loss of AtBMI1a and AtBMI1b retards seed germination and causes transcriptional derepression and a delayed chromatin state switch from H3K4me3 to H3K27me3 enrichment of several seed developmental genes (e.g. ABI3, DOG1, CRU3, CHO1). We found that AL6 and the PRC1 H3K27me3-reader component LHP1 directly bind at ABI3 and DOG1 loci. In light of these data, we propose that AL PHD-PRC1 complexes, built around H3K4me3, lead to a switch from the H3K4me3-associated active to the H3K27me3-associated repressive transcription state of seed developmental genes during seed germination. Our finding of physical interactions between PHD-domain proteins and PRC1 is striking and has important implications for understanding the connection between the two functionally opposite chromatin marks: H3K4me3 in activation and H3K27me3 in repression of gene transcription.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Structural Analysis of the Arabidopsis AL2-PAL and PRC1 Complex Provides Mechanistic Insight into Active-to-Repressive Chromatin State Switch.J Mol Biol. 2018 Oct 19;430(21):4245-4259. doi: 10.1016/j.jmb.2018.08.021. Epub 2018 Aug 31. J Mol Biol. 2018. PMID: 30176245
-
ZRF1 Chromatin Regulators Have Polycomb Silencing and Independent Roles in Development.Plant Physiol. 2016 Nov;172(3):1746-1759. doi: 10.1104/pp.16.00193. Epub 2016 Sep 14. Plant Physiol. 2016. PMID: 27630184 Free PMC article.
-
The putative PRC1 RING-finger protein AtRING1A regulates flowering through repressing MADS AFFECTING FLOWERING genes in Arabidopsis.Development. 2014 Mar;141(6):1303-12. doi: 10.1242/dev.104513. Epub 2014 Feb 19. Development. 2014. PMID: 24553292
-
Chromatin modulation and gene regulation in plants: insight about PRC1 function.Biochem Soc Trans. 2018 Aug 20;46(4):957-966. doi: 10.1042/BST20170576. Epub 2018 Jul 31. Biochem Soc Trans. 2018. PMID: 30065110 Review.
-
Governing the Silencing State of Chromatin: The Roles of Polycomb Repressive Complex 1 in Arabidopsis.Plant Cell Physiol. 2017 Feb 1;58(2):198-206. doi: 10.1093/pcp/pcw209. Plant Cell Physiol. 2017. PMID: 28069891 Review.
Cited by
-
The Histone Chaperone HIRA Is a Positive Regulator of Seed Germination.Int J Mol Sci. 2021 Apr 14;22(8):4031. doi: 10.3390/ijms22084031. Int J Mol Sci. 2021. PMID: 33919775 Free PMC article.
-
MORF-RELATED GENE702, a Reader Protein of Trimethylated Histone H3 Lysine 4 and Histone H3 Lysine 36, Is Involved in Brassinosteroid-Regulated Growth and Flowering Time Control in Rice.Plant Physiol. 2015 Aug;168(4):1275-85. doi: 10.1104/pp.114.255737. Epub 2015 Apr 8. Plant Physiol. 2015. PMID: 25855537 Free PMC article.
-
Characterizations of a Class-I BASIC PENTACYSTEINE Gene Reveal Conserved Roles in the Transcriptional Repression of Genes Involved in Seed Development.Curr Issues Mol Biol. 2022 Sep 7;44(9):4059-4069. doi: 10.3390/cimb44090278. Curr Issues Mol Biol. 2022. PMID: 36135190 Free PMC article.
-
The DAG1 transcription factor negatively regulates the seed-to-seedling transition in Arabidopsis acting on ABA and GA levels.BMC Plant Biol. 2016 Sep 9;16(1):198. doi: 10.1186/s12870-016-0890-5. BMC Plant Biol. 2016. PMID: 27613195 Free PMC article.
-
Family-Wide Characterization of Histone Binding Abilities of PHD Domains of AL Proteins in Arabidopsis thaliana.Protein J. 2018 Dec;37(6):531-538. doi: 10.1007/s10930-018-9796-4. Protein J. 2018. PMID: 30259302
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

