Differential effects of collagen prolyl 3-hydroxylation on skeletal tissues
- PMID: 24465224
- PMCID: PMC3900401
- DOI: 10.1371/journal.pgen.1004121
Differential effects of collagen prolyl 3-hydroxylation on skeletal tissues
Erratum in
- PLoS Genet. 2014 Jun;10(6):e1004473
Abstract
Mutations in the genes encoding cartilage associated protein (CRTAP) and prolyl 3-hydroxylase 1 (P3H1 encoded by LEPRE1) were the first identified causes of recessive Osteogenesis Imperfecta (OI). These proteins, together with cyclophilin B (encoded by PPIB), form a complex that 3-hydroxylates a single proline residue on the α1(I) chain (Pro986) and has cis/trans isomerase (PPIase) activity essential for proper collagen folding. Recent data suggest that prolyl 3-hydroxylation of Pro986 is not required for the structural stability of collagen; however, the absence of this post-translational modification may disrupt protein-protein interactions integral for proper collagen folding and lead to collagen over-modification. P3H1 and CRTAP stabilize each other and absence of one results in degradation of the other. Hence, hypomorphic or loss of function mutations of either gene cause loss of the whole complex and its associated functions. The relative contribution of losing this complex's 3-hydroxylation versus PPIase and collagen chaperone activities to the phenotype of recessive OI is unknown. To distinguish between these functions, we generated knock-in mice carrying a single amino acid substitution in the catalytic site of P3h1 (Lepre1(H662A) ). This substitution abolished P3h1 activity but retained ability to form a complex with Crtap and thus the collagen chaperone function. Knock-in mice showed absence of prolyl 3-hydroxylation at Pro986 of the α1(I) and α1(II) collagen chains but no significant over-modification at other collagen residues. They were normal in appearance, had no growth defects and normal cartilage growth plate histology but showed decreased trabecular bone mass. This new mouse model recapitulates elements of the bone phenotype of OI but not the cartilage and growth phenotypes caused by loss of the prolyl 3-hydroxylation complex. Our observations suggest differential tissue consequences due to selective inactivation of P3H1 hydroxylase activity versus complete ablation of the prolyl 3-hydroxylation complex.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




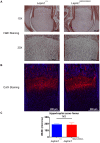

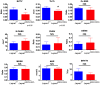
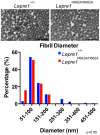

Similar articles
-
Null mutations in LEPRE1 and CRTAP cause severe recessive osteogenesis imperfecta.Cell Tissue Res. 2010 Jan;339(1):59-70. doi: 10.1007/s00441-009-0872-0. Epub 2009 Oct 28. Cell Tissue Res. 2010. PMID: 19862557 Free PMC article. Review.
-
Prolyl 3-hydroxylase 1 and CRTAP are mutually stabilizing in the endoplasmic reticulum collagen prolyl 3-hydroxylation complex.Hum Mol Genet. 2010 Jan 15;19(2):223-34. doi: 10.1093/hmg/ddp481. Epub 2009 Oct 21. Hum Mol Genet. 2010. PMID: 19846465 Free PMC article.
-
Mutations in PPIB (cyclophilin B) delay type I procollagen chain association and result in perinatal lethal to moderate osteogenesis imperfecta phenotypes.Hum Mol Genet. 2011 Apr 15;20(8):1595-609. doi: 10.1093/hmg/ddr037. Epub 2011 Jan 31. Hum Mol Genet. 2011. PMID: 21282188 Free PMC article.
-
Recessive osteogenesis imperfecta caused by LEPRE1 mutations: clinical documentation and identification of the splice form responsible for prolyl 3-hydroxylation.J Med Genet. 2009 Apr;46(4):233-41. doi: 10.1136/jmg.2008.062729. Epub 2008 Dec 16. J Med Genet. 2009. PMID: 19088120
-
Components of the collagen prolyl 3-hydroxylation complex are crucial for normal bone development.Cell Cycle. 2007 Jul 15;6(14):1675-81. doi: 10.4161/cc.6.14.4474. Epub 2007 May 18. Cell Cycle. 2007. PMID: 17630507 Review.
Cited by
-
An Update on Animal Models of Osteogenesis Imperfecta.Calcif Tissue Int. 2022 Oct;111(4):345-366. doi: 10.1007/s00223-022-00998-6. Epub 2022 Jun 29. Calcif Tissue Int. 2022. PMID: 35767009 Review.
-
Update on the Genetics of Osteogenesis Imperfecta.Calcif Tissue Int. 2024 Dec;115(6):891-914. doi: 10.1007/s00223-024-01266-5. Epub 2024 Aug 11. Calcif Tissue Int. 2024. PMID: 39127989 Free PMC article. Review.
-
Bone structure assessed by HR-pQCT, TBS and DXL in adult patients with different types of osteogenesis imperfecta.Osteoporos Int. 2015 Oct;26(10):2431-40. doi: 10.1007/s00198-015-3156-4. Epub 2015 May 9. Osteoporos Int. 2015. PMID: 25956285
-
Genetic analysis of osteogenesis imperfecta in the Palestinian population: molecular screening of 49 affected families.Mol Genet Genomic Med. 2018 Jan;6(1):15-26. doi: 10.1002/mgg3.331. Epub 2017 Nov 18. Mol Genet Genomic Med. 2018. PMID: 29150909 Free PMC article.
-
Bone involvement in adult patients affected with Ehlers-Danlos syndrome.Osteoporos Int. 2016 Aug;27(8):2525-31. doi: 10.1007/s00198-016-3562-2. Epub 2016 Apr 15. Osteoporos Int. 2016. PMID: 27084695
References
-
- Rauch F, Glorieux FH (2004) Osteogenesis imperfecta. Lancet 363: 1377–1385. - PubMed
-
- Morello R, Bertin TK, Chen Y, Hicks J, Tonachini L, et al. (2006) CRTAP is required for prolyl 3- hydroxylation and mutations cause recessive osteogenesis imperfecta. Cell 127: 291–304. - PubMed
-
- Tiainen P, Pasanen A, Sormunen R, Myllyharju J (2008) Characterization of recombinant human prolyl 3-hydroxylase isoenzyme 2, an enzyme modifying the basement membrane collagen IV. J Biol Chem 283: 19432–19439. - PubMed
-
- Vranka JA, Sakai LY, Bachinger HP (2004) Prolyl 3-hydroxylase 1: Enzyme characterization and identification of a novel family of enzymes. J Biol Chem 279: 23615–23621. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- F31 DE020954/DE/NIDCR NIH HHS/United States
- R01 AR036794/AR/NIAMS NIH HHS/United States
- P30 HD024064/HD/NICHD NIH HHS/United States
- HD22657/HD/NICHD NIH HHS/United States
- T32 GM008307/GM/NIGMS NIH HHS/United States
- DE01771/DE/NIDCR NIH HHS/United States
- P01 HD070394/HD/NICHD NIH HHS/United States
- R01 GM079656/GM/NIGMS NIH HHS/United States
- HD070394/HD/NICHD NIH HHS/United States
- R37 AR037318/AR/NIAMS NIH HHS/United States
- HD024064/HD/NICHD NIH HHS/United States
- P01 HD022657/HD/NICHD NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

