Interplay between Epigenetics and Genetics in Cancer
- PMID: 24465226
- PMCID: PMC3897842
- DOI: 10.5808/GI.2013.11.4.164
Interplay between Epigenetics and Genetics in Cancer
Abstract
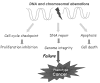
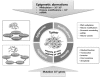
Genomic instability, which occurs through both genetic mechanisms (underlying inheritable phenotypic variations caused by DNA sequence-dependent alterations, such as mutation, deletion, insertion, inversion, translocation, and chromosomal aneuploidy) and epigenomic aberrations (underlying inheritable phenotypic variations caused by DNA sequence-independent alterations caused by a change of chromatin structure, such as DNA methylation and histone modifications), is known to promote tumorigenesis and tumor progression. Mechanisms involve both genomic instability and epigenomic aberrations that lose or gain the function of genes that impinge on tumor suppression/prevention or oncogenesis. Growing evidence points to an epigenome-wide disruption that involves large-scale DNA hypomethylation but specific hypermethylation of tumor suppressor genes, large blocks of aberrant histone modifications, and abnormal miRNA expression profile. Emerging molecular details regarding the modulation of these epigenetic events in cancer are used to illustrate the alterations of epigenetic molecules, and their consequent malfunctions could contribute to cancer biology. More recently, intriguing evidence supporting that genetic and epigenetic mechanisms are not separate events in cancer has been emerging; they intertwine and take advantage of each other during tumorigenesis. In addition, we discuss the collusion between epigenetics and genetics mediated by heterochromatin protein 1, a major component of heterochromatin, in order to maintain genome integrity.
Keywords: epigenomics; genetics; heterochromatin-specific nonhistone chromosomal protein HP-1; neoplasms.
Figures



References
-
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. - PubMed
-
- Waddington CH. Towards a theoretical biology. Nature. 1968;218:525–527. - PubMed
-
- Waddington CH. The epigenotype. 1942. Int J Epidemiol. 2012;41:10–13. - PubMed
-
- Wu C, Morris JR. Genes, genetics, and epigenetics: a correspondence. Science. 2001;293:1103–1105. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
