Phylogenetic and biological significance of evolutionary elements from metazoan mitochondrial genomes
- PMID: 24465405
- PMCID: PMC3896360
- DOI: 10.1371/journal.pone.0084330
Phylogenetic and biological significance of evolutionary elements from metazoan mitochondrial genomes
Abstract
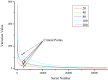
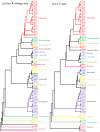
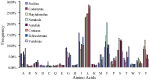
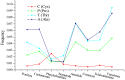
The evolutionary history of living species is usually inferred through the phylogenetic analysis of molecular and morphological information using various mathematical models. New challenges in phylogenetic analysis are centered mostly on the search for accurate and efficient methods to handle the huge amounts of sequence data generated from newer genome sequencing. The next major challenge is the determination of relationships between the evolution of structural elements and their functional implementation, which is largely ignored in previous analyses. Here, we described the discovery of structural elements in metazoan mitochondrial genomes, termed key K-strings, that can serve as a basis for phylogenetic tree construction. Although comprising only a small fraction (0.73%) of all K-strings, these key K-strings are pivotal to the tree construction because they allow for a significant reduction in the computational time required to construct phylogenetic trees, and more importantly, they make significant improvement to the results of phylogenetic inference. The trees constructed from the key K-strings were consistent overall to our current view of metazoan phylogeny and exhibited a more rational topology than the trees constructed by using other conventional methods. Surprisingly, the key K-strings tended to accumulate in the conserved regions of the original sequences, which were most likely due to strong selection pressure. Furthermore, the special structural features of the key K-strings should have some potential applications in the study of the structures and functions relationship of proteins and in the determination of evolutionary trajectory of species. The novelty and potential importance of key K-strings lead us to believe that they are essential evolutionary elements. As such, they may play important roles in the process of species evolution and their physical existence. Further studies could lead to discoveries regarding the relationship between evolution and processes of speciation.
Conflict of interest statement
Figures
Similar articles
-
Complete mitochondrial genomes reveal phylogeny relationship and evolutionary history of the family Felidae.Genet Mol Res. 2013 Sep 3;12(3):3256-62. doi: 10.4238/2013.September.3.1. Genet Mol Res. 2013. PMID: 24065666
-
Neogastropod phylogenetic relationships based on entire mitochondrial genomes.BMC Evol Biol. 2009 Aug 23;9:210. doi: 10.1186/1471-2148-9-210. BMC Evol Biol. 2009. PMID: 19698157 Free PMC article.
-
Phylogeny and evolution of Cervidae based on complete mitochondrial genomes.Genet Mol Res. 2012 Mar 14;11(1):628-35. doi: 10.4238/2012.March.14.6. Genet Mol Res. 2012. PMID: 22535398
-
Complete mitochondrial genomes of edible mushrooms: features, evolution, and phylogeny.Physiol Plant. 2024 May-Jun;176(3):e14363. doi: 10.1111/ppl.14363. Physiol Plant. 2024. PMID: 38837786 Review.
-
Genetic aspects of mitochondrial genome evolution.Mol Phylogenet Evol. 2013 Nov;69(2):328-38. doi: 10.1016/j.ympev.2012.10.020. Epub 2012 Nov 7. Mol Phylogenet Evol. 2013. PMID: 23142697 Review.
Cited by
-
Resolution of deep divergence of club fungi (phylum Basidiomycota).Synth Syst Biotechnol. 2019 Dec 10;4(4):225-231. doi: 10.1016/j.synbio.2019.12.001. eCollection 2019 Dec. Synth Syst Biotechnol. 2019. PMID: 31890927 Free PMC article.
-
Phylogenomic proximity and comparative proteomic analysis of SARS-CoV-2.Gene Rep. 2020 Sep;20:100777. doi: 10.1016/j.genrep.2020.100777. Epub 2020 Jul 8. Gene Rep. 2020. PMID: 32835133 Free PMC article.
-
CVTree: A Parallel Alignment-free Phylogeny and Taxonomy Tool Based on Composition Vectors of Genomes.Genomics Proteomics Bioinformatics. 2021 Aug;19(4):662-667. doi: 10.1016/j.gpb.2021.03.006. Epub 2021 Jun 10. Genomics Proteomics Bioinformatics. 2021. PMID: 34119695 Free PMC article.
-
Computational approach for the identification of putative allergens from Cucurbitaceae family members.J Food Sci Technol. 2021 Jan;58(1):267-280. doi: 10.1007/s13197-020-04539-7. Epub 2020 May 26. J Food Sci Technol. 2021. PMID: 33505071 Free PMC article.
-
Temperature-Driven Intraspecific Diversity in Paralytic Shellfish Toxin Profiles of the Dinoflagellate Alexandrium pacificum and Intragenic Variation in the Saxitoxin Biosynthetic Gene, sxtA4.Microb Ecol. 2025 Aug 8;88(1):87. doi: 10.1007/s00248-025-02586-1. Microb Ecol. 2025. PMID: 40779244 Free PMC article.
References
-
- Stenderup JT, Olesen J, Glenner H (2006) Molecular phylogeny of the Branchiopoda (Crustacea)—multiple approaches suggest a ‘diplostracan’ ancestry of the Notostraca. Mol Phylogenet Evol 41: 182–194. - PubMed
-
- Teichmann SA, Mitchison G (1999) Making family trees from gene families. Nat Genet 21: 66–67. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

