Development of ultra-high-density screening tools for microbial "omics"
- PMID: 24465499
- PMCID: PMC3897414
- DOI: 10.1371/journal.pone.0085177
Development of ultra-high-density screening tools for microbial "omics"
Abstract
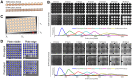
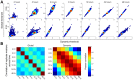
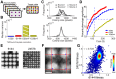
High-throughput genetic screens in model microbial organisms are a primary means of interrogating biological systems. In numerous cases, such screens have identified the genes that underlie a particular phenotype or a set of gene-gene, gene-environment or protein-protein interactions, which are then used to construct highly informative network maps for biological research. However, the potential test space of genes, proteins, or interactions is typically much larger than current screening systems can address. To push the limits of screening technology, we developed an ultra-high-density, 6144-colony arraying system and analysis toolbox. Using budding yeast as a benchmark, we find that these tools boost genetic screening throughput 4-fold and yield significant cost and time reductions at quality levels equal to or better than current methods. Thus, the new ultra-high-density screening tools enable researchers to significantly increase the size and scope of their genetic screens.
Conflict of interest statement
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

