Genome-wide transcriptome and antioxidant analyses on gamma-irradiated phases of deinococcus radiodurans R1
- PMID: 24465634
- PMCID: PMC3900439
- DOI: 10.1371/journal.pone.0085649
Genome-wide transcriptome and antioxidant analyses on gamma-irradiated phases of deinococcus radiodurans R1
Abstract
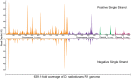
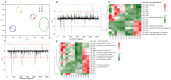
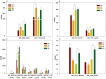
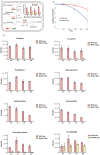
Adaptation of D. radiodurans cells to extreme irradiation environments requires dynamic interactions between gene expression and metabolic regulatory networks, but studies typically address only a single layer of regulation during the recovery period after irradiation. Dynamic transcriptome analysis of D. radiodurans cells using strand-specific RNA sequencing (ssRNA-seq), combined with LC-MS based metabolite analysis, allowed an estimate of the immediate expression pattern of genes and antioxidants in response to irradiation. Transcriptome dynamics were examined in cells by ssRNA-seq covering its predicted genes. Of the 144 non-coding RNAs that were annotated, 49 of these were transfer RNAs and 95 were putative novel antisense RNAs. Genes differentially expressed during irradiation and recovery included those involved in DNA repair, degradation of damaged proteins and tricarboxylic acid (TCA) cycle metabolism. The knockout mutant crtB (phytoene synthase gene) was unable to produce carotenoids, and exhibited a decreased survival rate after irradiation, suggesting a role for these pigments in radiation resistance. Network components identified in this study, including repair and metabolic genes and antioxidants, provided new insights into the complex mechanism of radiation resistance in D. radiodurans.
Conflict of interest statement
Figures


References
-
- Hagen U (1994) Mechanisms of induction and repair of DNA double-strand breaks by ionizing radiation: some contradictions. Radiat Environ Biophys 33: 45–61. - PubMed
-
- Daly MJ, Gaidamakova E, Matrosova V, Vasilenko A, Zhai M, et al. (2004) Accumulation of Mn (II) in Deinococcus radiodurans facilitates gamma-radiation resistance. Science 306: 1025–1028. - PubMed
-
- Cox MM, Battista JR (2005) Deinococcus radiodurans - the consummate survivor. Nat Rev Microbiol 3: 882–892. - PubMed
-
- Pocker Y, Li H (1991) Kinetics and mechanism of methanol and formaldehyde interconversion and formaldehyde oxidation catalyzed by liver alcohol dehydrogenase. Adv Exp Med Biol 284: 315–325. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

