Signal intensities derived from different NMR probes and parameters contribute to variations in quantification of metabolites
- PMID: 24465670
- PMCID: PMC3897511
- DOI: 10.1371/journal.pone.0085732
Signal intensities derived from different NMR probes and parameters contribute to variations in quantification of metabolites
Erratum in
- PLoS One. 2014;9(7):e102929
Abstract
We discovered that serious issues could arise that may complicate interpretation of metabolomic data when identical samples are analyzed at more than one NMR facility, or using slightly different NMR parameters on the same instrument. This is important because cross-center validation metabolomics studies are essential for the reliable application of metabolomics to clinical biomarker discovery. To test the reproducibility of quantified metabolite data at multiple sites, technical replicates of urine samples were assayed by 1D-(1)H-NMR at the University of Alberta and the University of Michigan. Urine samples were obtained from healthy controls under a standard operating procedure for collection and processing. Subsequent analysis using standard statistical techniques revealed that quantitative data across sites can be achieved, but also that previously unrecognized NMR parameter differences can dramatically and widely perturb results. We present here a confirmed validation of NMR analysis at two sites, and report the range and magnitude that common NMR parameters involved in solvent suppression can have on quantitated metabolomics data. Specifically, saturation power levels greatly influenced peak height intensities in a frequency-dependent manner for a number of metabolites, which markedly impacted the quantification of metabolites. We also investigated other NMR parameters to determine their effects on further quantitative accuracy and precision. Collectively, these findings highlight the importance of and need for consistent use of NMR parameter settings within and across centers in order to generate reliable, reproducible quantified NMR metabolomics data.
Conflict of interest statement
Figures
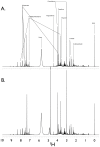


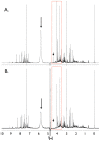
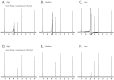
References
-
- Nicholson JK, Lindon JC (2008) Systems biology: Metabonomics. Nature 455: 1054–1056. - PubMed
-
- Dunn WB, Broadhurst DI, Atherton HJ, Goodacre R, Griffin JL (2011) Systems level studies of mammalian metabolomes: the roles of mass spectrometry and nuclear magnetic resonance spectroscopy. Chem Soc Rev 40: 387–426. - PubMed
-
- Foxall PJ, Parkinson JA, Sadler IH, Lindon JC, Nicholson JK (1993) Analysis of biological fluids using 600 MHz proton NMR spectroscopy: application of homonuclear two-dimensional J-resolved spectroscopy to urine and blood plasma for spectral simplification and assignment. J Pharm Biomed Anal 11: 21–31. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

