Characterization and interactome study of white spot syndrome virus envelope protein VP11
- PMID: 24465701
- PMCID: PMC3897518
- DOI: 10.1371/journal.pone.0085779
Characterization and interactome study of white spot syndrome virus envelope protein VP11
Abstract
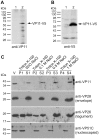
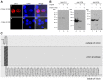
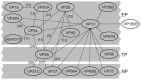
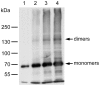
White spot syndrome virus (WSSV) is a large enveloped virus. The WSSV viral particle consists of three structural layers that surround its core DNA: an outer envelope, a tegument and a nucleocapsid. Here we characterize the WSSV structural protein VP11 (WSSV394, GenBank accession number AF440570), and use an interactome approach to analyze the possible associations between this protein and an array of other WSSV and host proteins. Temporal transcription analysis showed that vp11 is an early gene. Western blot hybridization of the intact viral particles and fractionation of the viral components, and immunoelectron microscopy showed that VP11 is an envelope protein. Membrane topology software predicted VP11 to be a type of transmembrane protein with a highly hydrophobic transmembrane domain at its N-terminal. Based on an immunofluorescence assay performed on VP11-transfected Sf9 cells and a trypsin digestion analysis of the virion, we conclude that, contrary to topology software prediction, the C-terminal of this protein is in fact inside the virion. Yeast two-hybrid screening combined with co-immunoprecipitation assays found that VP11 directly interacted with at least 12 other WSSV structural proteins as well as itself. An oligomerization assay further showed that VP11 could form dimers. VP11 is also the first reported WSSV structural protein to interact with the major nucleocapsid protein VP664.
Conflict of interest statement
Figures




Similar articles
-
Characterization of white spot syndrome virus envelope protein VP51A and its interaction with viral tegument protein VP26.J Virol. 2008 Dec;82(24):12555-64. doi: 10.1128/JVI.01238-08. Epub 2008 Oct 1. J Virol. 2008. PMID: 18829765 Free PMC article.
-
A 3D model of the membrane protein complex formed by the white spot syndrome virus structural proteins.PLoS One. 2010 May 19;5(5):e10718. doi: 10.1371/journal.pone.0010718. PLoS One. 2010. PMID: 20502662 Free PMC article.
-
Identification of the nucleocapsid, tegument, and envelope proteins of the shrimp white spot syndrome virus virion.J Virol. 2006 Mar;80(6):3021-9. doi: 10.1128/JVI.80.6.3021-3029.2006. J Virol. 2006. PMID: 16501111 Free PMC article.
-
VP26 of white spot syndrome virus functions as a linker protein between the envelope and nucleocapsid of virions by binding with VP51.J Virol. 2008 Dec;82(24):12598-601. doi: 10.1128/JVI.01732-08. Epub 2008 Oct 8. J Virol. 2008. PMID: 18842736 Free PMC article.
-
Characterization of a novel envelope protein WSV010 of shrimp white spot syndrome virus and its interaction with a major viral structural protein VP24.Virology. 2007 Jul 20;364(1):208-13. doi: 10.1016/j.virol.2007.02.030. Epub 2007 Apr 2. Virology. 2007. PMID: 17400271
Cited by
-
Laminin Receptor in Shrimp Is a Cellular Attachment Receptor for White Spot Syndrome Virus.PLoS One. 2016 Jun 3;11(6):e0156375. doi: 10.1371/journal.pone.0156375. eCollection 2016. PLoS One. 2016. PMID: 27257954 Free PMC article.
-
Crystal Structure of Major Envelope Protein VP24 from White Spot Syndrome Virus.Sci Rep. 2016 Aug 30;6:32309. doi: 10.1038/srep32309. Sci Rep. 2016. PMID: 27572278 Free PMC article.
-
Structural modelling and preventive strategy targeting of WSSV hub proteins to combat viral infection in shrimp Penaeus monodon.PLoS One. 2024 Jul 29;19(7):e0307976. doi: 10.1371/journal.pone.0307976. eCollection 2024. PLoS One. 2024. PMID: 39074084 Free PMC article.
-
A VP24-truncated isolate of white spot syndrome virus is inefficient in per os infection.Vet Res. 2017 Dec 11;48(1):87. doi: 10.1186/s13567-017-0492-8. Vet Res. 2017. PMID: 29228988 Free PMC article.
References
-
- Vlak JM, Bonami JR, Flegel TW, Kou G-H, Lightner DV, et al.. (2005) Nimaviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, eds. VIIIth Report of the International Committee on Taxonomy of Viruses, Elsevier. 187–192.
-
- Escobedo-Bonilla CM, Alday-Sanz V, Wille M, Sorgeloos P, Pensaert MB, et al. (2008) A review on the morphology, molecular characterization, morphogenesis and pathogenesis of white spot syndrome virus. J Fish Dis 31: 1–18. - PubMed
-
- Lo C-F, Peng S-E, Chang Y-S, Kou G-H (2005) White spot syndrome− What we have learned about the virus and the disease. In: Walker PJ, Lester RG, Bondad-Reantaso MG, eds. Diseases in Asian Aquaculture V. Proceedings of the 5th Symposium on Diseases in Asian Aquaculture. Fish Health Section, Asian Fisheries Society, Manila. 421–433.
-
- Wang C-H, Lo C-F, Leu J-H, Chou C-M, Yeh P-Y, et al. (1995) Purification and genomic analysis of baculovirus associated with white spot syndrome (WSBV) of Penaeus monodon. . Dis Aquat Organ 23: 239–242.
-
- Li Z, Lin Q, Chen J, Wu JL, Lim TK, et al. (2007) Shotgun identification of the structural proteome of shrimp white spot syndrome virus and iTRAQ differentiation of envelope and nucleocapsid subproteomes. Mol Cell Proteomics 6: 1609–1620. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

